Translate this page into:
Spectrum of Virulence Factors in Clinical Isolates of Staphylococcus aureus and Prevalence of SCCmec Types in Methicillin-Resistant Staphylococcus aureus in a Tertiary Care Center
Address for correspondence: Shanthi Mariappan, MBBS, MD, PhD, Professor, Department of Microbiology, Sri Ramachandra Institute of Higher Education and Research (SRIHER), Porur, Chennai 600 116, Tamil Nadu, India (e-mail: shanthi.m@sriramachandra.edu.in).
-
Received: ,
Accepted: ,
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is a widely recognized multidrug-resistant bacteria presenting a major therapeutic challenge to clinicians. Staphylococcus aureus possesses a number of pathogenicity factors that attribute to the severity of infections. This study was undertaken to investigate the common virulence genes in clinical isolates of Staphylococcus aureus, determine their antimicrobial susceptibility profile, and to characterize the staphylococcal cassette chromosome mec (SCCmec) types among MRSA in a tertiary care center.
Materials and Methods
A total of 133 clinical isolates were included in this study. Susceptibility to various antibiotics was determined by disc diffusion method. Methicillin resistance was screened using cefoxitin disc; mecA and mecC genes were detected using polymerase chain reaction (PCR). PCR was done to detect 12 virulence factors such as hla, hlb, fnbA, fnbB, sea, seb, sec, icaA, clfA, tst, pvl, and eta. SCCmec typing was done by multiplex PCR.
Results
Of the 133 clinical isolates, 54 (40.6%) were MRSA. The most common virulence gene detected was hlb (61.6%), hla (39%), and fnbA (37%). SCCmec type I was the most predominant. Mortality rate of 6.7% was observed among patients with staphylococcal infections. Univariate analysis of mortality associated virulence genes did not reveal any significant association between virulence genes and mortality.
Conclusion
The distribution of virulence genes is similar in both MRSA and methicillin-sensitive Staphylococcus aureus. MRSA belongs to the SCCmec types I to IV. Possession of multiple virulence factors and multidrug resistance profile makes Staphylococcus aureus a formidable pathogen in clinical settings.
Keywords
methicillin-resistant Staphylococcus aureus
MRSA
Staphylococcus aureus
virulence genes
SCCmec typing
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a public health concern on a global scale and a widely recognized multidrug-resistant bacteria.[1] In many countries, 25 to 50% of all staphylococcal infections are attributable to MRSA.[2] Though it is an essential component of the human microbiota, it is associated with a multitude of infections including skin and soft tissue, bacteremia, pneumonia, osteomyelitis, endocarditis, and toxin-mediated diseases.[3]
Most MRSA possesses the mecA-encoded, low affinity, penicillin binding protein A movable genetic element found in the chromosome of MRSA called the staphylococcal cassette chromosome mec (SCCmec) and harbors the mecA and/or mecC genes, mediating methicillin resistance.[4,5]
Staphylococcus aureus possesses virulence determinants genes that help the organism survive and colonize or interfere with the host defense system. They include various toxins like leucocidins, enterotoxins, exfoliative toxins, exotoxins, and hemolysis and surface components like capsules, peptidoglycans, teichoic acid, and protein A, in addition to enzymes like esterase, lipases, fatty acid modifying enzymes, proteases, hydrolytic enzymes, and catalases.[1] Biofilm production is another crucial factor contributing to therapeutic resistance and pathogenicity. Multidrug resistance and the combination of the above pathogenic factors attribute to severity of infections.[3] The surface proteins of Staphylococcus aureus aids in adhesion and colonization. The cellular proteins, proteases, and toxins prevent phagocytosis and blunt the immune response.[6]
Enterotoxin genes, sea, seb, and sec, are associated with food poisoning. The serine protease enzyme such as exfoliative toxin encoded by the gene eta is related to blister development, cell- to-cell adhesion, and keratinocyte loss. The tst gene encodes for toxic shock syndrome toxin, an exoprotein and a member of the pyrogenic toxin super antigen family, and impairs the immunological response of the cells, ultimately causing cell death.[6]
Staphylococcus aureus produces exotoxins[7] that are cytolytic which form β-barrel pores in the plasma membrane, leading to leakage of the cell contents and lysis cytolysins include α-hemolysin, β-hemolysin, γ-hemolysin, leukocidin, and Panton-Valentine leukocidin (PVL).[8] The hla gene codes for the α-hemolysin that is lethal to human epithelial cells and a primary agent in skin infections. The beta-hemolysin, encoded by the hlb gene, is a sphingomyelinase and is presumed to have a role in lung injury and pneumonia.[9]
Panton Valentine leukocidin (pvl) is a synergohymenotropic toxin that also causes pores in cell membranes. Although sepsis and necrotizing pneumonia have been documented in association with pvl-producing MRSA, most often they cause minor skin or soft tissue infections. Pvl is primarily encountered in community-associated MRSA and infrequently in-hospital isolates; thus, it is regarded as a marker for community-acquired strains. It exhibits affinity towards leukocytes.[7]
Staphylococcal scalded skin syndrome is linked to the, exfoliative toxins namely, ETA and ETB that are pyrogenic exoproteins. The exfoliative toxins, ETA and ETB, are encoded by the eta and etb genes, respectively. Despite sharing the same biological functions, ETA and ETB have different immunological traits. The eta gene is considered to occur in higher frequency.[10]
The intercellular adhesion cluster, which contains the ica gene, aids in the formation of biofilm. The clumping factors, clfA and clfB, fibronectin-binding proteins A (FnBA) and B (FnBB), collagen binding protein, and staphylococcal protein A are typical members of microbial surface component recognizing adhesive matrix molecules and are critical for adherence.[7] The fnbA and fnbB favor cell internalization and intercellular persistence leading to chronic infections.[6] Clumping factor A (ClfA) is linked to the fibrinogen region causing bacterial or platelet aggregation in the blood.[11]
Rich genetic diversity in the SCCmec element causes proliferation of virulence and antibiotic resistance determinants. Till date, 15 SCCmec types (I–XV) have been recognized, based on the combinations of five mec complexes (classes A, B, C1, C2, and E) and nine ccr gene complexes (types I–IX).[12] The variety of SCCmec types and virulence factors creates a reservoir for transmission of infections.
This study was undertaken to investigate the common virulence genes in clinical isolates of Staphylococcus aureus, determine their antimicrobial susceptibility profile, and to characterize the SCCmec types among MRSA in a tertiary-care teaching hospital.
Materials and Methods
Bacterial Isolates
The study was conducted in a 1,600-bed university teaching hospital in South India from 2019 to 2021. A total of 133 clinically significant, consecutive, nonrepetitive Staphylococcus aureus isolated from patients hospitalized for more than 48 hours were included in the study. Repetitive isolates from the same patients and isolates from outpatients were excluded from the study. The study protocol was approved by Institutional Ethics Committee (REF: IEC-NI/19/FEB/68/12).
The source of the isolates was exudates (n = 105), blood (n = 18), urine (n = 7), and respiratory specimens (n = 3). The significance of the study isolates was ascertained by correlating with Gram stain, significant growth in cultures, and the patient clinical history. The isolates were identified up to species level by standard biochemical tests and automated systems: VITEK2 GP-card (bioMerieux, Marcy l'Etoile, France) and MALDI-TOF MS (bioMerieux, Marcy l'Etoile, France).
Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing was done by Kirby Bauer disc diffusion for different classes of antimicrobial agents such as ampicillin (10µg), cefuroxime (30µg), erythromycin (30µg), clindamycin (2µg), amikacin (30µg), ciprofloxacin (5µg), and linezolid (30µg). Methicillin resistance was detected by using cefoxitin (30µg) disc (Himedia, Mumbai, Maharashtra, India) as per Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI-M100-S29).[13] Minimum inhibitory concentrations of linezolid (MicroExpress, Goa, India) teicoplanin and vancomycin were determined by agar dilution method in accordance with CLSI guidelines.
Phenotypic Methods
Production of Biofilm: Microtiter Plate Method
Production of biofilm was determined by microtiter plate method using 96-well polystyrene microtiter plate in method described previously.[14] The Staphylococcus aureus was classified as strong biofilm producers with OD of more than 0.240, moderate biofilm producers with optical density OD of 0.120 to 0.240, and nonbiofilm producers with OD of less than 0.120.
Molecular Methods
DNA Extraction
DNA extraction was performed by boiling-lysis method as described previously.[15] Colonies of clinical strains were transferred to 1000 µL of sterile distilled water. The samples were then boiled to prepare the DNA. It was centrifuged at 1,200rpm for 10 minutes. The supernatant was separated and stored. This was used as template for polymerase chain reactions (PCRs).
Polymerase Chain Reaction
All the isolates were subjected to molecular confirmation using the species specific nuc gene.[16]MecA and mecC genes were amplified to detect methicillin resistance.[17,18] Triplex, duplex, and simplex PCR were set up to detect the genes encoding virulence factors such as icaA (intracellular adhesin protein), hla and hlb (hemolysin toxin alpha and hemolysin toxin beta), pvl (Panton valentine toxin), sea, seb and sec (staphylococcal enterotoxin A, staphylococcal enterotoxin B, and staphylococcal enterotoxin c), eta (exfoliative toxin A), clfA (clumping factor), tst (toxic shock syndrome), fnbA and fnbB (fibronectin binding protein), with the following PCR conditions: Initial denaturation at 94°C for 5 minutes, denaturation at 94°C for 15 seconds, annealing: variable for 30 seconds, and extension at 72°C for 35 cycles for 30 seconds, Final extension at 72°C for 45 seconds. All the PCR reactions were carried out with a final volume of 25 µL reaction. Each reaction contained 10pmol of each primer (Eurofins, Bangalore, India) and 23 µL of master mix (Takara, India) and 2µL of template DNA.[19–21]
Multiplex PCR was set up for SCCmec typing with the following PCR conditions: Initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 35 cycles for 1 minute, final extension at 72°C for 7 minutes. PCR was performed in a mixture of 30 μL volume containing 15 μL master mix (Takara, India); 0.5 μL of each of βF1, α3R1, ccrCF, and ccrCR primers (10pmol/μL; Eurofins, India); 0.3μL of each of 1272F1, 1272R1, 5RmecAF, 5R431R primers (10pmol/μL; Eurofins, India; and 2μL template DNA. The final volume was adjusted to 30μL by adding 9.8μL sterile ultrapure water (Takara, India).[22]
The amplicons were separated in a 2% agarose gel containing ethidium bromide (►Fig. 1). The primers used are described in ►Table 1. Previously, characterized strains were used as positive controls. Sterile Mili Q water was used as negative controls.
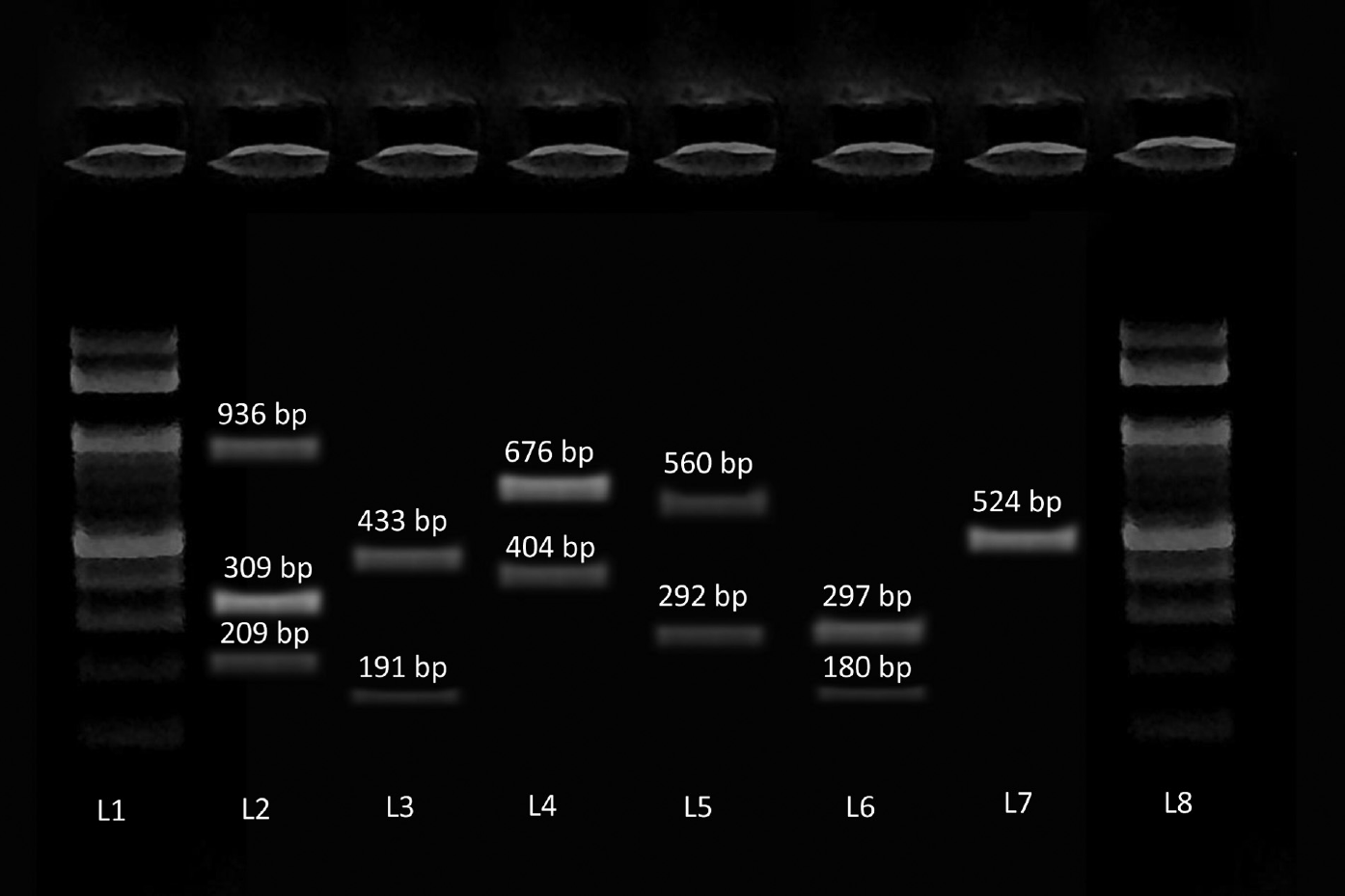
- Image of gel electrophoresis of polymerase chain reaction for the detection of virulence genes of Staphylococcus aureus. DNA ladder (100bp) (L1 and L8), icaA, hla and hlb (L2), pvl and fnbA (L3), eta and seb (L4), sea and clfA (L5), sec and tst (L6), fnbB (L7).
| Gene | Primer sequence | Temperature | Amplicon | Reference | |
|---|---|---|---|---|---|
| Methicillin resistance | nuc | F- TCGCTTGCTATGATTGTGG R- GCCAATGTTCTACCATAGC |
50 | 359 | 15 |
| mecA | F- GTAGAAATGACTGAACGTCCGATAA R- CCAATTCCACATTGTTTCGGTCTAA |
55 | 310 | 16 | |
| mecC | F- GAAAAAAAGGCTTAGAACGCCTC R- GAAGATCTTTTCCGTTTTCAGC |
54 | 132 | 17 | |
| Virulence genes | icaA | F- TCAGACACTTGCTGGCGCAGTC R- TCACGATTCTCTCCCTCTCTGCCATT |
56 | 936 | 18 |
| hlb | F- GTGCACTTACTGACAATAGTGC R- GTTGATGAGTAGCTACCTTCAGT |
309 | 19 | ||
| hla | F- CTGATTACTATCCAAGAAATTCGATTG R- CTTTCCAGCCTACTTTTTTATCAGT |
209 | |||
| eta | F- CGCTGCGGACATTCCAACATGG R- TACATGCCCGCCACTTGCTTGT |
60 | 676 | ||
| seb | F- ATTCTATTAAGGACACTAAGTTAGGGA R- ATCCCGTTTCATAAGGCGAGT |
404 | |||
| sea | F- GAAAAAAGTCTGAATTGCAGGGAACA R- CAAATAAATCGTAATTAACCGAAGGTTC |
56 | 676 | ||
| clfA | F- ATTGGCGTGGCTTCAGTGCT R- CGTTTCTTCCGTAGTTGCATTTG |
292 | |||
| sec | F- GTAAAGTTACAGGTGGCAAAACTTG R- CATATCATACCAAAAAGTATTGCCGT |
57 | 297 | ||
| tst | F- TTCACTATTTGTAAAAGTGTCAGACCCACT R- TACTAATGAATTTTTTTATCGTAAGCCCTT |
180 | |||
| fnbB | F- GTAACAGCTAATGGTCGAATTGATACT R- CAAGTTCGATAGGAGTACTATGTTC |
58 | 524 | ||
| pvl | F- ATCATTAGGTAAAATGTCTGGACATGATCC R- GCATCAASTGTATTGGATAGCAAAAGC |
58 | 433 | ||
| fnbA | F- GATACAAACCCAGGTGGTGG R- TGTGCTTGACCATGYTCTTC |
191 | 20 | ||
| SCCmec typing | β | F- ATTGCCTTGATAATAGCCYTCT R- TAAAGGCATCAATGCACAAACACT |
55 | 937 | 21 |
| a3 | |||||
| ccrC | F- CGTCTATTACAAGATGTTAAGGATAAT R- CCTTTATAGACTGGATTATTCAAAATAT |
518 | |||
| ccrC | |||||
| 1272 | F- GCCACTCATAACATATGGAA R- CATCCGAGTGAAACCCAAA |
415 | |||
| 1272 | |||||
| 5RmecA | F- TATACCAAACCCGACAACTAC R- CGGCTACAGTGATAACATCC |
359 | |||
| 5R431 |
Abbreviations: PCR, polymerase chain reaction; SCCmec, staphylococcal cassette chromosome mec.
The obtained sequences were submitted in the GenBank database under the following accession numbers: ON500665, ON500666, ON500667, ON500668, ON500669, ON500670, ON500671, ON500672, OP125544, OP125545.
Statistical Analysis
Statistical analysis was done by using SPSS software 16.0 version (IBM Corp., New York, United States). Chi-squared test and Fisher's exact test was used to compare the distribution of virulence genes among MRSA and methicillin-sensitive Staphylococcus aureus(MSSA) isolates. Univariate analysis was done to find the significance between virulence factors and mortality. The p-value of less than 0.05 was considered as statistically significant.
Results
Antimicrobial Susceptibility Pattern
The resistance exhibited to various classes of antimicrobials is as follows: ampicillin 73.6% (98/133), cefuroxime 40.6% (54/133), cefoxitin 40.6% (54/133), erythromycin 48.1% (64/133), clindamycin 27% (36/133), and ciprofloxacin 61.6 % (82/133). All isolates were susceptible to linezolid, vancomycin and teicoplanin.
Phenotypic Methods
Biofilm Production by Microtiter Plate Method
Forty percent (53/133) of the study isolates were biofilm producers by microtiter plate method that included 7.5% (4/53) strong biofilm producers and 92.5% (49/53) moderate biofilm producers. The remaining 60% (80/133) were nonbiofilm producers.
Molecular Methods
Among the 133 isolates, the nuc gene was present in all the isolates, confirming the identification as Staphylococcus aureus.
Detection of Methicillin Resistance
Of the 133 study isolates, 54 (40.6%) were MRSA and carried the mecA gene. MecC gene was not detected in any of the study isolates.
Virulence Genes Profile
In this study, 81.3% (108/133) isolates of Staphylococcus aureus carried one or more virulence genes. The most common virulence gene detected in this study was hlb 61.6% (82/133) followed by hla 39% (52/133) and fnbA 37.5% (50/133). The detection of the virulence gene among the total study isolates and their distribution among MRSA and MSSA is depicted in ►Table 2. Twenty-five (18.7%) study isolates did not harbor any of the genes that were looked for in this study.
| Virulence genes | Staphylococcus aureus (n = 133) | Percentage | MRSA (n = 54) | Percentage | MSSA (n = 79) | Percentage | p-Value |
|---|---|---|---|---|---|---|---|
| icaA | 46 | 34.5 | 18 | 33.3 | 28 | 35.2 | 0.80166 |
| hla | 52 | 39 | 23 | 42.5 | 29 | 36.7 | 0.49468 |
| hlb | 82 | 61.6 | 32 | 59.2 | 50 | 63.2 | 0.63862 |
| pvl | 9 | 6.7 | 5 | 9.2 | 4 | 5 | 0.4847 |
| fnbA | 50 | 37.5 | 21 | 38.8 | 29 | 36.7 | 0.79879 |
| fnbB | 20 | 15 | 6 | 11.1 | 14 | 17.7 | 0.29491 |
| sea | 28 | 21 | 10 | 18.5 | 18 | 22.7 | 0.55340 |
| seb | 19 | 14.2 | 6 | 11.1 | 13 | 16.4 | 0.38703 |
| sec | 10 | 7.5 | 4 | 7.4 | 6 | 7.5 | 0.96787 |
| clfA | 20 | 15 | 9 | 16.6 | 11 | 13.9 | 0.66388 |
| eta | 9 | 6.7 | 3 | 5.5 | 6 | 7.5 | 0.7381 |
| tst | 14 | 10.5 | 6 | 11.1 | 8 | 10.1 | 0.85582 |
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus.
In 15.7% (17/108) of the isolates, only a single virulence gene was detected. The virulence genes that occurred singly were sea (4), hla (3), hlb (3), clfA (2), icaA (2), seb (1), sec (1), and tst (1).
Of the 108 isolates that carried the virulence genes, 91 (84.3%) carried more than one gene in various combinations. Some combinations of virulence genes were found in single isolates only. These combinations are depicted in ►Tables 3 and 4, respectively.
| Virulence genes | Number of isolates (n = 48) | MRSA (18) | MSSA (30) |
|---|---|---|---|
| icaA, hla, hlb, fnbB | 3 (6.25%) | 1 | 2 |
| hla, hlb, fnbA, sea | 3 (6.25%) | 1 | 2 |
| hlb, eta | 3 (6.25%) | 1 | 2 |
| sec, tst | 3 (6.25%) | 1 | 2 |
| hla, fnbA | 2 (4.1%) | 2 | 0 |
| hla, hlb | 2 (4.1%) | 0 | 2 |
| hla, hlb, fnbA | 2 (4.1%) | 0 | 2 |
| hla, hlb, fnbA, seb | 2 (4.1%) | 1 | 1 |
| hla, hlb, sea | 2 (4.1%) | 1 | 1 |
| hla, hlb, sea, seb | 2 (4.1%) | 1 | 1 |
| hlb, fnbA | 2 (4.1%) | 2 | 0 |
| hlb, fnbB | 2 (4.1%) | 0 | 2 |
| hlb, tst | 2 (4.1%) | 0 | 2 |
| icaA, hla, hlb | 2 (4.1%) | 1 | 1 |
| icaA, hla, hlb, fnbA, clfA, fnbB | 2 (4.1%) | 0 | 2 |
| icaA, hla, hlb, fnbA, sea, clfA, seb | 2 (4.1%) | 1 | 1 |
| icaA, hla, hlb, fnbA, tst | 2 (4.1%) | 1 | 1 |
| icaA, hla, hlb, fnbA | 2 (4.1%) | 1 | 1 |
| icaA, hlb | 2 (4.1%) | 1 | 1 |
| icaA, hlb, fnbA, sec | 2 (4.1%) | 1 | 1 |
| icaA, hlb, fnbB | 2 (4.1%) | 1 | 1 |
| sea, seb | 2 (4.1%) | 0 | 2 |
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus.
| Virulence genes in single isolates (n = 43) | MRSA (18) | MSSA (25) |
|---|---|---|
| fnbA, sea, fnbB | 0 | 1 |
| hla, clfA | 0 | 1 |
| hla, hlb, fnbA, clfA | 1 | 0 |
| hla, hlb, fnbA, clfA, eta | 0 | 1 |
| hla, hlb, fnbA, eta | 1 | 0 |
| hla, hlb, sea, clfA | 0 | 1 |
| hla, hlb, fnbB | 0 | 1 |
| hla, hlb, sea, tst | 1 | 0 |
| hla, sea, sec | 0 | 1 |
| hlb, fnbA, clfA | 0 | 1 |
| hlb, fnbA, fnbB | 0 | 1 |
| hlb, fnbA, seb | 1 | 0 |
| hlb, pvl, fnbA, seb | 0 | 1 |
| hlb, pvl, fnbB, clfA, eta, seb | 0 | 1 |
| hlb, sec | 1 | 0 |
| hlb, sec, tst | 1 | 0 |
| icaA, hla, fnbA | 0 | 1 |
| icaA, hla, fnbB | 0 | 1 |
| icaA, hla, hlb, clfA, sea | 1 | 0 |
| icaA, hla, hlb, fnbA, clfA, fnbB, eta | 1 | 0 |
| icaA, hla, hlb, fnbA, clfA, seb | 1 | 0 |
| icaA, hla, hlb, fnbA, fnbB, eta | 0 | 1 |
| icaA, hla, hlb, fnbA, sea, fnbB | 1 | 0 |
| icaA, hla, hlb, fnbA, seb | 0 | 1 |
| icaA, hla, hlb, fnbA, sea, seb | 0 | 1 |
| icaA, hla, hlb, pvl, fnbA, clfA | 1 | 0 |
| icaA, hla, hlb, pvl, fnbA, fnbB | 0 | 1 |
| icaA, hla, hlb, pvl, fnbA, sea, clfA | 1 | 0 |
| icaA, hla, hlb, pvl, fnbA, sec, tst | 0 | 1 |
| icaA, hla, hlb, pvl, fnbA, tst | 1 | 0 |
| icaA, hla, hlb, pvl, sea, seb | 0 | 1 |
| icaA, hla, hlb, sea | 1 | 0 |
| icaA, hla, hlb, sea, seb | 0 | 1 |
| icaA, hlb, clfA, fnbB | 0 | 1 |
| icaA, hlb, fnbA, clfA, seb | 0 | 1 |
| icaA, hlb, fnbA, sea, seb | 0 | 1 |
| icaA, hlb, fnbA, seb | 1 | 0 |
| icaA, hlb, sea | 1 | 0 |
| icaA, hlb, tst | 0 | 1 |
| icaA, hla, hlb, pvl, fnbA | 1 | 0 |
| icaA, seb | 0 | 1 |
| pvl, fnbA, fnbB | 1 | 0 |
| sea, fnbB, eta | 0 | 1 |
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus.
Among the exudative isolate, the most common gene was hlb followed by hla and fnbA. The pvl gene was detected only in exudative isolates. The most common gene among the blood isolates was hlb. The respiratory isolates carried the hlb and icaA gene. ►Table 5 depicts the distribution of the virulence gene of Staphylococcus aureus in various clinical specimens.
| Virulence genes | Exudates (105) | Blood (18) | Urine (7) | Respiratory (3) |
|---|---|---|---|---|
| icaA | 37 (35.2%) | 5 (27.7%) | 1 (14.2%) | 3 (100%) |
| hlb | 68 (64.7%) | 7 (38.8%) | 4 (57.1%) | 3 (100%) |
| hla | 42 (40%) | 5 (27.7%) | 2 (28.5%) | 2 (66.7%) |
| pvl | 9 (8.5%) | 0 | 0 | 0 |
| fnbA | 43 (40.9%) | 4 (22.2%) | 2 (28.5%) | 1 (33.4%) |
| eta | 8 (7.6%) | 1 (5.5%) | 0 | 0 |
| seb | 15 (14.2%) | 2 (11.1%) | 1 (14.2%) | 0 |
| sea | 21 (20%) | 3 (16.6%) | 4 (57.1%) | 0 |
| clfA | 17 (16.1%) | 2 (11.1%) | 1 (14.2%) | 0 |
| sec | 8 (7.6%) | 2 (11.1%) | 0 | 0 |
| tst | 11 (10.4%) | 2 (11.1%) | 0 | 1 (33.4%) |
| fnbB | 14 (13.3%) | 4 (22.2%) | 1 (14.2%) | 1 (33.4%) |
SCCmec Typing
By SCCmec typing, among the 54 MRSA isolates, 64.8% (35/54) were typable. Nineteen (35.1%) isolates were nontypable. Of the 35 typable MRSA, 26 belonged to SCCmec type I, 7 to type IV, and one belonged to type II. One isolate belonged to both the type I and III. The antimicrobial susceptibility profile of the SCCmec types is depicted in ►Table 6.
| Antibiotics | SCCmec Type I (n = 26) | SCCmec type II (n = 1) | SCCmec type IV (n = 7) | SCCmec type I and III (n = 1) | Non typable (n = 19) |
|---|---|---|---|---|---|
| Erythromycin | 8 (30.7%) | 0 | 3 (42.8%) | 0 | 11 (57.8%) |
| Clindamycin | 15 (57.6%) | 0 | 4 (57.1%) | 1 (100%) | 16 (84.2%) |
| Ciprofloxacin | 5 (19.2%) | 0 | 4 (57.1%) | 0 | 6 (31.5%) |
| Gentamycin | 7 (26.9%) | 1 (100%) | 4 (57.1%) | 0 | 12 (63.1%) |
| Teicoplanin | 26 (100%) | 1 (100%) | 7 (100%) | 1 (100%) | 19 (100%) |
| Linezolid | 26 (100%) | 1 (100%) | 7 (100%) | 1 (100%) | 19 (100%) |
| Vancomycin | 26 (100%) | 1 (100%) | 7 (100%) | 1 (100%) | 19 (100%) |
Abbreviation: SCCmec, staphylococcal cassette chromosome mec.
Statistical Analysis
A mortality rate of 6.7% (9/133) was observed among the patients with staphylococcal infection that included six MSSA and three MRSA. All the three MRSAs belonged to SCCmec type I. The virulence gene profile, underlying disease condition in these patients and the antibiotics susceptibility profile, is given in ►Table 7. Univariate analysis of mortality associated virulence gene did not reveal any significant association between mortality and the presence virulence genes (►Table 8).
| Specimens | Virulence genes | Diagnosis | MRSA/MSSA | Antibiotic susceptibility profile |
|---|---|---|---|---|
| Exudate | icaA, hla, hlb, fnbA, sea, seb | Sepsis, DM, multiple organ failure | MSSA | Clindamycin Cloxacillin Cefuroxime Linezolid Vancomycin Teicoplanin |
| Blood | sec, tst | Pneumonia, Sepsis | MSSA | Ampicillin Cloxacillin Erythromycin Clindamycin Ciprofloxacin Gentamycin Cefuroxime Linezolid Vancomycin Teicoplanin |
| Blood | hla, clfA | CKD, DM, CRBSI | MSSA | Cloxacillin Erythromycin Clindamycin Ciprofloxacin Cefuroxime Linezolid Vancomycin Teicoplanin |
| Blood | hlb, fnbB | Diabetic foot, Sepsis | MSSA | Ampicillin Cloxacillin Cefuroxime Linezolid Vancomycin Teicoplanin |
| Exudate | icaA, hlb, fnbA, clfA, seb | Sepsis, CKD, DM | MSSA | Ampicillin Cefuroxime Cloxacillin Erythromycin Clindamycin Gentamycin Linezolid Vancomycin Teicoplanin |
| Blood | icaA, hlb, fnbA, sea, seb | Urosepsis. DM | MSSA | Cloxacillin Clindamycin Ciprofloxacin Gentamycin Cefuroxime Linezolid Vancomycin Teicoplanin |
| Blood | icaA, hla, hlb, fnbB | Diabetic foot ulcer, Sepsis | MRSA (SCCmec type I) |
Erythromycin Clindamycin Ciprofloxacin Gentamycin Linezolid Vancomycin Teicoplanin |
| Exudate | None | CKD, Diabetic cellulitis, Sepsis | MRSA (SCCmec type I) |
Linezolid Vancomycin Teicoplanin |
| Blood | icaA, hlb, hla, sea | Sepsis, AKI, DM, CAD | MRSA (SCCmec type I) |
Erythromycin Clindamycin Ciprofloxacin Linezolid Vancomycin Teicoplanin |
Abbreviations: AKI, acute kidney injury; CAD, coronary artery disease; CKD, chronic kidney disease; CRBSI, catheter-related bloodstream infection; DM, diabetes mellitus; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; SCCmec, staphylococcal cassette chromosome mec.
| Virulence genes | Deceased (n = 9) | Survivors (n = 124) | Odd's ratio 95% confidence interval | p-Value |
|---|---|---|---|---|
| icaA | 2.5305 (0.6449–9.9256) | 0.1831 | ||
| Present | 5 | 41 | ||
| Absent | 4 | 83 | ||
| hla | 1.2667 (0.3239–4.9525) | 0.7340 | ||
| Present | 4 | 48 | ||
| Absent | 5 | 76 | ||
| hlb | 1.2632 (0.3016–5.2903) | 0.7492 | ||
| Present | 6 | 76 | ||
| Absent | 3 | 48 | ||
| pvl | 0.6399 (0.0345–11.8620) | 0.7644 | ||
| Present | 0 | 9 | ||
| Absent | 9 | 115 | ||
| fnbA | 0.7872 (0.1878–3.3000) | 0.7436 | ||
| Present | 3 | 47 | ||
| Absent | 6 | 77 | ||
| eta | 0.6399 (0.0345–11.8620) | 0.7644 | ||
| Present | 0 | 9 | ||
| Absent | 9 | 115 | ||
| seb | 3.3750 (0.7667–14.8566) | 0.1077 | ||
| Present | 3 | 16 | ||
| Absent | 6 | 108 | ||
| sea | 1.9800 (0.4627–8.4723) | 0.3570 | ||
| Present | 3 | 25 | ||
| Absent | 6 | 99 | ||
| clfA | 1.6825 (0.3234–8.7524) | 0.5363 | ||
| Present | 2 | 18 | ||
| Absent | 7 | 106 | ||
| sec | 1.5972 (0.1793–14.2258) | 0.6747 | ||
| Present | 1 | 9 | ||
| Absent | 8 | 115 | ||
| tst | 1.0673 (0.1235–9.2255) | 0.9528 | ||
| Present | 1 | 13 | ||
| Absent | 8 | 111 | ||
| fnbB | 1.6825 (0.3234–8.7524) | 0.5363 | ||
| Present | 2 | 18 | ||
| Absent | 7 | 106 |
Discussion
There has been an increase in mortality and morbidity due to Staphylococcus aureus infections, both in the community and in health care settings. The pathogenicity of Staphylococcus aureus is due to the presence of various virulence factors that undermine the immune system and promote their survival and persistence.
Of the 133 study isolates, 54 (40.6%) isolates were MRSA. The prevalence of MRSA in a recent multicentric study conducted in India ranged from 32% to 80%.[23] According to the Antimicrobial Resistance Surveillance network (ICMR), the prevalence of MRSA in the Indian population ranged from 21% to 45%.[24] A study conducted in North India over a period of 3 years reported that MRSA increased from 28% in 2017 to 35.1% in 2019 with overall prevalence being 33.7%.[25] The prevalence of MRSA infection was 37.34% according to a recent meta-analysis report from South India.[26] In Asia-Pacific region, MRSA accounts for up to 50% of Staphylococcus aureus bloodstream infections. In North Europe, South Europe, Australia, and the United States, prevalence of MRSA was less than 5%, 25% to 50%, 20% to 33%, and 53%, respectively.[27]
In this study, virulence genes in both MRSA and MSSA were investigated. Presence of one or more virulence genes was observed in 81.3% (108/133) of the study isolates. In both staphylococcal biotypes, various combinations of virulence genes were present. According to few studies, some genes are known to occur more frequently in each of the biotypes.[28] Twenty-five (18.7%) isolates did not harbor any of the genes that were looked for in this study.
The genes encoding for fnbA and fnbB were encountered in 37.5% and 15% isolates, respectively, in this study. fnbA was the most common that is in concordance with studies from China and Palestine.[20,29] However, in both these reports, prevalence of fnbA was higher than this study with 78.2% and 62.5%, respectively. Occurrence of fnbB was 29% in both the above reports.
In the current investigation, 15% of the isolates carried the clfA gene. There is a marked variation in the detection of clfA gene in clinical isolates of Staphylococcus aureus. In China, a study reported that all their study isolates carried the clfA gene, while a study from Brazil did not detect the clfA gene in any isolate.[3,20] In Indian study, 58.7% MRSA harbored the clfA gene.[30]
Polysaccharide intercellular adhesin, which is required for biofilm development, is encoded for and controlled by the intercellular adhesion (icaADCB) operon. Among the ica genes, icaA has been shown to be crucial for Staphylococcus aureus for the development of biofilms. The N-acetylglucosaminyltransferase enzyme, which generates N-acetylglucosamine oligomers from UDP-N-acetylglucosamine, is encoded by the icaA gene. In this study, icaA was detected in 34.5% (46/133) isolates, of which majority were from exudative 82.6% (38/46) isolates followed by blood 10.8% (5/46). Biofilm production was observed by microtiter plate in 39.8% (53/133) of the study isolates. Of these biofilm producers, icaA was present in 19 isolates only. The remaining 34 biofilm producers lacked icaA, indicating the presence of other biofilm encoding genes namely, icaB, icaC, and icaD, that were not included in the study protocol. The detection of icaA gene in this study was lower when compared to a study from North India, wherein 52.3% harbored the icaA gene.[31] Studies from Brazil, China, and Iran reported 100%, 89.9%, and 60.3% of icaA, respectively. Hence, icaA may not be the sole gene associated with biofilm formation.[32,33]
The hla and hlb genes that encode for cytolytic toxins such as alpha hemolysin and beta hemolysin in Staphylococcus aureus were the most common virulence genes in this study. Majority of the isolates (91.7%, 122/133) in the present investigation carried at least one of the hemolysin encoding gene, hla and/or hlb. Eleven isolates did not harbor any of the hemolysins. Hlb was more common than hla with their occurrence being 61.6% and 39%, respectively. In Iran also hlb was the most common hemolysin observed, while in China hla was the most predominant.[1,20]
In this study among the enterotoxin encoding genes, sea gene was the most common and was present in 21% of the isolates followed by seb and sec with occurrence of 14.2 and 7.5%, respectively. This result is in agreement with an investigation from Iran who reported 40.6%, 19.6% and 3% of the sea, seb, and sec.[34] Three studies from China observed that, among the enterotoxin genes, seb occurred more frequently.[2,8,20] In Brazil, the investigators were able to detect sea (3.23%) and seb (54.84%) genes, respectively, but failed to detect the sec gene.[3]
In this study, the occurrence of eta and tst genes was 5.2 and 10.5%, respectively. Previous studies have found wide variation (0.2% to 94.4%) in the occurrence of the eta gene in Staphylococcus aureus.[8,10,20,34–36] The prevalence of tst gene ranged between 5% and 34.6% in various published reports.[8,32,35,37–40]
Pvl gene was detected in 6.7% (9/133) and all were from isolates recovered from wound exudates. The occurrence of pvl gene is low, when compared to that of countries in the West.[41] Other investigators from other countries have found the prevalence of pvl gene to be ranging from 11.3% to 42.8%.[2,3,8,20,36,40,42]
In this study, the adhesion (fnbA, fnbB, and clfA) and cytolysins (hlb and hla) genes occurred more frequently particularly among isolates from exudative specimens. This suggests that adherence and cytotoxicity are majorly operative in skin and skin and soft tissue infections by Staphylococcus aureus
In this study, there is no significant difference in the distribution of virulence genes among MRSA and MSSA. This is in consensus to a study from Brazil that investigated 13 virulence genes in Staphylococcus aureus and found that there was no association between the presence of the virulence genes and methicillin resistance.[40] However, some studies have found significant variation. An investigator from Columbia inferred that the virulence genes were more diverse and frequent in MSSA than in MRSA (83 vs. 73%).[43] On the contrary, in Nigeria, a study compared the distribution of eight virulence genes among MRSA and MSSA and found that there was a significant association between MRSA and the presence of two virulence genes, eta and icaA.[28] Another study from Hungary found that, out of the 14 studied virulence genes, adhesins like cna, sea, ica, and hlb were significantly more in MRSA, whereas superantigens like tst, eta, and sec were significantly abundant in MSSA.[44] The results of our study demonstrate that MSSA strains continue to be a significant cause of infection, indicating that MRSA has not completely replaced MSSA strains in clinical settings.
It has been frequently demonstrated that hospital-associated MRSA carried SCCmec types I, II, and III, while community-acquired MRSA belonged to types IV and V.[22] This characteristic distribution of SCCmec types is becoming increasingly irrelevant as evidenced by the detection of different SCCmec types in hospital settings. According to many published reports both within India and other countries, the prevalence of different SCCmec types of MRSA strains varied considerably. In this study, of the 54 MRSA, 26 (48.1%) belonged to SCCmec type I followed by 12.9% (7/54) and 1.8% (1/54) to type IV and II, respectively. SCCmec type V was not found in any of the study isolates. One isolate was of dual type (type I and III). Presence of two SCCmec types in a single isolate has been described, in studies from Sikkim and Hyderabad (1.1–3.3%).[45,46] Notably in Canada the majority of the isolates were of dual type (73.58%).[47] A proportion of our isolates were nontypeable (n = 19/54, 35.2%). In previous reports from India, the frequency of nontypeable MRSA was only 4% to 7.6%. The recent report from Hyderabad also found that 33.3% of the MRSA were not typeable in their study and this was attributed to the presence of clones endemic to the region and have suggested that large-scale multicentric investigation from the South Asian region would be required to gain further insights.[45] In this study, the antibiotic susceptibility profile also did not reveal any notable difference among the various SCCmec types investigated in this study (►Table 6).
In this study, all the MRSA belonging to SCCmec types I, II, and IV carried different combinations of virulence genes investigated. This is in concordance with reports from India, Nigeria, Hungary, and Korea, wherein the MRSA isolates belonging to the different SCCmec types carried one or more virulence genes.[6,28,44,48,49,50] In our study, one isolate that belonged to both the type I and III did not carry any of the virulence determinants. This may have harbored other virulence genes not included in the study protocol. In this study, of the nine pvl gene carriers and five were MRSA. Among these five pvl positive MRSA, two each belonged to SCCmec type I and IV and one belonged to type II.
In this study, the mortality rate was 6.76% (9/133) among the patients with staphylococcal infection that include six MSSA and three MRSA. While eight isolates from these patients carried one or more virulence genes, one isolate did not carry any of the virulence gene investigated in the study. However, the mortality rate was lower than studies from United states and Cape Town which reported that the in-hospital mortality attributed to Staphylococcus aureus infection was 13% and 29%, respectively.[51,52]
In this study, by univariate analysis, it was evident that none of the virulence genes had significant association with mortality. This was similar to the observation in a study from Hungary which reported that the number of virulence genes carried and the presence of specific virulence factors did not influence the outcome of infection even though they had a reported a high mortality rate of 35.3% among patients with staphylococcal infections.[44]
The characterization of multiple virulence genes in clinical isolates and their correlation with clinical outcomes can be considered as the strength of this study. Such a type of study has not been undertaken in India and elsewhere. A case-controlled study with large number of clinical isolates can provide more insights into the association of virulence factors with morbidity and mortality.
Conclusions
The distribution of virulence genes is similar in both MRSA and MSSA. MRSA belongs to the SCCmec types I to IV. In view of the nontypeable MRSA encountered in the study, the other SCCmec types, VI to XV, have to be investigated. Though the prevalence of MRSA is 40.6%, the possession of multiple virulence factors and multidrug resistance profile makes MRSA a formidable pathogen in clinical settings.
Conflict of Interest
None declared.
Funding
This study was supported by the Founder chancellor Shri N.P.V. Ramasamy Udayar fellowship, provided by Sri Ramachandra Institute of Higher Education and Research.
References
- Virulence factors of Methicillin Resistant Staphylococcus aureus (MRSA) isolated from burn patients. Int J Curr Microbiol Appl Sci. 2015;4(07):898-906.
- [Google Scholar]
- Antimicrobial resistance, virulence genes profiling and molecular relatedness of methicillin-resistant Staphylococcus aureus strains isolated from hospitalized patients in Guangdong Province, China. Infect Drug Resist. 2019;12:447-459.
- [CrossRef] [PubMed] [Google Scholar]
- Virulence factors in methicillin-resistant Staphylococcus aureus isolated from ICU units in Brazil. Adv Microbiol. 2014;4:207-215.
- [CrossRef] [Google Scholar]
- Current status of staphylococcal cassette chromosome mec (SCCmec) Antibiotics (Basel). 2022;11(01):86.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical and molecular epidemiology of staphylococcal toxic shock syndrome in the United Kingdom. Emerg Infect Dis. 2018;24(02):258-266.
- [CrossRef] [Google Scholar]
- Distribution of virulence genes and SCCmec types among methicillin-resistant Staphylococcus aureus of clinical and environmental origin: a study from community of Assam, India. BMC Res Notes. 2021;14(01):58.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of virulence factors of Staphylococcus aureus: novel function of known virulence factors that are implicated in activation of airway epithelial proinflammatory response. J Pathogens. 2011;2011:601905.
- [CrossRef] [PubMed] [Google Scholar]
- Clonality, virulence genes, and antibiotic resistance of Staphylococcus aureus isolated from blood in Shandong, China. BMC Microbiol. 2021;21(01):281.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of hemolysins of Staphylococcus strains isolated from human and bovine, southern Iran. Majallah-i Tahqiqat-i Dampizishki-i Iran. 2014;15(04):326-330.
- [Google Scholar]
- Prevalence of exfoliative toxin A and B genes in Staphylococcus aureus isolated from clinical specimens. J Infect Public Health. 2014;7(03):177-185.
- [CrossRef] [PubMed] [Google Scholar]
- Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog. 2011;7(10):e1002307.
- [CrossRef] [PubMed] [Google Scholar]
- Novel SCCmec type XV (7A) and two pseudo-SCCmec variants in foodborne MRSA in China. J Antimicrob Chemother. 2022;77(04):903-909.
- [CrossRef] [PubMed] [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI supplement (29th). Wayne, PA: CLSI; 2019.
- [Google Scholar]
- Detection of biofilm producing staphylococci among different clinical isolates and its relation to methicillin susceptibility. Open Access Maced J Med Sci. 2018;6(08):1335-1341.
- [CrossRef] [PubMed] [Google Scholar]
- Analysis of antibiotic resistance genes and its associated SCCmec types among nasal carriage of methicillin resistant coagulase negative staphylococci from community settings, Chennai, Southern India. J Clin Diagn Res. 2015;9(08):DC01-DC05.
- [CrossRef] [PubMed] [Google Scholar]
- Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J Clin Microbiol. 2011;49(10):3627-3631.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence and molecular characterization of methicillin resistance among Coagulase-negative Staphylococci at a tertiary care center. Med J Armed Forces India. 2016;72(Suppl. 01):S54-S58.
- [CrossRef] [PubMed] [Google Scholar]
- Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251) Clin Microbiol Infect. 2012;18(04):395-400.
- [CrossRef] [PubMed] [Google Scholar]
- Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J Clin Microbiol. 2008;46(02):678-684.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characteristics and virulence gene profiles of Staphylococcus aureus causing bloodstream infection. Braz J Infect Dis. 2018;22(06):487-494.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of cna, fnbA and fnbB adhesin genes among Staphylococcus aureus isolates from orthopedic infections associated to different types of implant. FEMS Microbiol Lett. 2005;246(01):81-86.
- [CrossRef] [PubMed] [Google Scholar]
- New update on molecular diversity of clinical Staphylococcus aureus isolates in Iran: antimicrobial resistance, adhesion and virulence factors, biofilm formation and SCCmec typing. Mol Biol Rep. 2022;49(04):3099-3111.
- [CrossRef] [PubMed] [Google Scholar]
- Treatment of MRSA infections in India: clinical insights from a Delphi analysis. Indian J Med Microbiol. 2022;40(01):35-45.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence and genetic mechanisms of antimicrobial resistance in Staphylococcus species: a multicentre report of the Indian Council of Medical Research antimicrobial resistance surveillance network. Indian J Med Microbiol. 2017;35(01):53-60.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence pattern of MRSA from a rural medical college of North India: a cause of concern. J Family Med Prim Care. 2021;10(02):752-757.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of Methicillin-Resistant Staphylococcus Aureus (MRSA) in India: A Systematic Review and Meta-Analysis.
- [Google Scholar]
- Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4 18033
- [CrossRef] [PubMed] [Google Scholar]
- Association of virulence genes with mecA gene in Staphylococcus aureus isolates from Tertiary Hospitals in Nigeria. Indian J Pathol Microbiol. 2015;58(04):464-471.
- [CrossRef] [PubMed] [Google Scholar]
- Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genomics. 2019;20(01):578.
- [CrossRef] [PubMed] [Google Scholar]
- In vitro and in vivo biofilm characterization of methicillin-resistant Staphylococcus aureus from patients associated with pharyngitis infection. BioMed Res Int. 2016;2016 1289157
- [CrossRef] [PubMed] [Google Scholar]
- Phenotypic detection of biofilm formation in clinical isolates of Methicillin-resistant Staphylococcus aureus. Asian Journal of Pharmaceutical Research and Health Care. 2022;14(01):43-47.
- [CrossRef] [Google Scholar]
- Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn Microbiol Infect Dis. 2012;74(04):363-368.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of icaABCD genes and biofilm formation in clinical isolates of methicillin resistant Staphylococcus aureus. Iran J Pathol. 2014;9(04):257-262.
- [Google Scholar]
- Virulence factors of Staphylococcus aureus isolates in an Iranian referral children's hospital. Osong Public Health Res Perspect. 2014;5(02):96-100.
- [CrossRef] [PubMed] [Google Scholar]
- Analysis of virulence genes among methicillin resistant Staphylococcus aureus (MRSA) strains. Jundishapur J Microbiol. 2014;7(06):e10741.
- [CrossRef] [PubMed] [Google Scholar]
- Virulence factor genes and antimicrobial susceptibility of Staphylococcus aureus strains isolated from blood and chronic wounds. Toxins (Basel). 2021;13(07):491.
- [CrossRef] [PubMed] [Google Scholar]
- Coexistence of virulence genes in methicillin-resistant Staphylococcus aureus clinical isolates. Rev Soc Bras Med Trop. 2018;51(03):361-363.
- [CrossRef] [PubMed] [Google Scholar]
- Antibiotic resistance patterns and virulence determinants of different SCCmec and pulsotypes of Staphylococcus aureus isolated from a major hospital in Ilam, Iran. Open Microbiol J. 2017;11:211-223.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol. 2003;41(04):1434-1439.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular epidemiology of methicillin-resistant Staphylococcus aureus in the Brazilian primary health care system. Trop Med Int Health. 2019;24(03):339-347.
- [CrossRef] [PubMed] [Google Scholar]
- A population-based study examining the emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 in New York City. Ann Clin Microbiol Antimicrob. 2006;5(01):29.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular and virulence characteristics of methicillin-resistant Staphylococcus aureus in burn patients. Frontiers in Laboratory Medicine. 2017;1(01):43-47.
- [CrossRef] [Google Scholar]
- Characterisation of virulence genes in methicillin susceptible and resistant Staphylococcus aureus isolates from a paediatric population in a university hospital of Medellín, Colombia. Mem Inst Oswaldo Cruz. 2011;106(08):980-985.
- [CrossRef] [PubMed] [Google Scholar]
- Characterisation of antibiotic resistance, virulence, clonality and mortality in MRSA and MSSA bloodstream infections at a tertiary-level hospital in Hungary: a 6-year retrospective study. Ann Clin Microbiol Antimicrob. 2020;19(01):17.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of community- & hospital-acquired methicillin-resistant & methicillin-sensitive Staphylococcus aureus isolates in Sikkim. Indian J Med Res. 2015;142(03):330-335.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterisation of methicillin-resistant Staphylococcus aureus isolated from patients at a tertiary care hospital in Hyderabad, South India. Indian J Med Microbiol. 2020;38(02):183-191.
- [CrossRef] [PubMed] [Google Scholar]
- Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43(10):5026-5033.
- [CrossRef] [PubMed] [Google Scholar]
- Antibiotic susceptibility, virulence pattern, and typing of Staphylococcus aureus strains isolated from variety of infections in India. Front Microbiol. 2019;10 2763
- [CrossRef] [PubMed] [Google Scholar]
- Clonal clusters and virulence factors of methicillin-resistant Staphylococcus Aureus: evidence for community-acquired methicillin-resistant Staphylococcus Aureus infiltration into hospital settings in Chennai, South India. Indian J Med Microbiol. 2019;37(03):326-336.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic relationship between SCCmec types and virulence factors of methicillin-resistant Staphylococcus aureus clinical isolates in Korea. J Exp Biomed Sci. 2010;16:75-78.
- [Google Scholar]
- Methicillin-susceptible and methicillin-resistant Staphylococcus aureus bacteremia: nationwide estimates of 30-day readmission, in-hospital mortality, length of stay, and cost in the United States. Clin Infect Dis. 2019;69(12):2112-2118.
- [CrossRef] [PubMed] [Google Scholar]
- The association between pathogen factors and clinical outcomes in patients with Staphylococcus aureus bacteraemia in a tertiary hospital, Cape Town. Int J Infect Dis. 2020;91:111-118.
- [CrossRef] [PubMed] [Google Scholar]





