Translate this page into:
Bloodstream infections and trends of antimicrobial sensitivity patterns at Port Blair
Address for correspondence: Dr. Amit Banik, Room# 203, AIIHPH, BN Campus, Salt Lake, Kolkata, West Bengal, India. E-mail: dramitbanik@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
PURPOSE:
Bloodstream infection can range from inapparent bacteremia until fulminant septic shock with high mortality. Microorganisms present in circulating blood whether continuously, intermittently, or transiently are a threat to every organ in the body. Culture of blood is a vital tool to diagnose such infections. Drug susceptibility patterns help in rationalizing therapy.
OBJECTIVE:
The objective of this study was to perform bacteriological analysis and assess drug sensitivity patterns of isolates from blood stream infections.
DESIGN:
Retrospective observational study was conducted from May 2015 to February 2017 at a tertiary care hospital, Port Blair, India. Blood samples were collected with aseptic guidelines and cultured for 7 days. Growths were identified using standard biochemical tests and subjected to sensitivity testing according to Modified Kirby–Bauer's disk diffusion method. Data for the source of blood collection and duration of incubation were noted and compared.
RESULTS:
A total of 270 (14.24%) pathogens were isolated from 1895 bacteremia suspect patient blood specimens. Contamination was observed at a rate of 1.63%. Gram-positive cocci (60.37%) were predominant organisms recovered followed by Gram-negative Bacilli (36.29%) and Yeasts (3.33%). Staphylococcus aureus, CoNS, and Acinetobacter spp. were the primary pathogens isolated. Aminoglycosides, carbapenems, and glycopeptides were the most effective drugs for treating bacteremia.
CONCLUSIONS:
Successful treatment of sepsis depends on early diagnosis and proper antimicrobial therapy. Local knowledge of bacteriological profile and antimicrobial sensitivity patterns helps rationalize empiric treatment strategies.
Keywords
Andaman and Nicobar Islands
antimicrobial sensitivity
blood culture
bloodstream infection
empiric therapy
Introduction
Blood is a connective tissue which is indispensable for proper functioning and survival of human life. From providing nutrients, limiting/clearing pathogens, perfusing and ventilating organs, clotting wounds, removal of toxins and chemicals, dissemination of hormones and drugs throughout the body it performs a pivotal role in body defense and survival. Blood is considered to be a sterile fluid unless proved otherwise. Invasion of the bloodstream by microorganisms constitutes one of the most serious situations in infectious disease. Microorganisms present in circulating blood whether continuously, intermittently, or transiently are a threat to every organ in the body.[1] Clinical presentation ranges from benign transient bacteremia with little or no symptoms to fulminant septic shock with high mortality.[2] Bloodstream infections (BSIs) have serious consequences such as shock, disseminated intravascular coagulation, multiple organ failure, and even death. Early diagnosis plays a crucial role in managing BSI, and hence, prompt detection of such infections is a critical function of clinical microbiology laboratories. Blood culture is a vital tool for the detection of BSI and remains the gold standard for bacteremia detection. Empiric antimicrobial therapy is based on knowledge of the microbial profile and their antimicrobial sensitivity patterns, clinical and epidemiological data. Irrational use of drugs has led to an increase of multidrug-resistant bugs and thus worsened the condition.[1] Prevalence and susceptibility patterns of microorganism vary according to geography and even within the same hospital with time. Andaman and Nicobar Islands (A and N islands) are a group of islands in Ritchie's Archipelago situated far away from the Indian mainland which bear strikingly different topography, climatic conditions, and a plethora of different ethnicities living together. With the background of no such information from these islands; the current study intends to report the prevalence, bacteriological analysis of microorganisms and antimicrobial susceptibility profiles of blood culture isolates from the main referral hospital at Port Blair in the A and N islands.
Materials and Methods
The present study is a retrospective observational analysis of blood culture isolates received in the Department of Microbiology, G B Pant Hospital attached to Andaman & Nicobar Islands Institute of Medical Sciences, Port Blair, from May 2015 to February 2017. Records of all such cases were perused from the medical records section of the hospital and analyzed for inclusion in the study.
Inclusion and exclusion criteria
Records of patients of all age groups with fever (both high- and low-grade) due to infective causes were included in the study. Patients with a history of having autoimmune or chronic diseases such as tuberculosis or sarcoidosis, immunodeficient patients, patients on steroids, having heat stroke or having a suspected viral or parasitic infection were excluded from the study. Consecutive, patient-specific, and nonduplicate isolates were included for analysis.
Sample collection
Blood specimens were obtained according to the sample collection protocol of hospital at the bedside by nursing staff, resident doctors from wards, critical care units, or by a trained phlebotomist. The protocol included that the skin be disinfected with 2 % Chlorhexidine, before drawing blood. The antecubital and median cubital fossa were the preferred sampling sites. The blood samples from the central vein catheters, if any were obtained from needleless caps that have been disinfected with 70% isopropyl alcohol, allowed to dry and wiped with sterile gauze or cotton before obtaining the sample.
Sample processing
Blood for culture was collected from 1895 clinically suspected bacteremia cases under strict aseptic precautions. A volume of 5–10 ml from adults and 2–3 ml from pediatric patients were obtained for culture. The same was inoculated into conventional pediatric and adult blood culture bottles containing brain–heart infusion broths (1:10 dilution). These were then continuously incubated aerobically at 37°C. After 18–24 h of incubation, a blind subculture was done to appropriate solid culture media irrespective of the turbidity status. The bottles were taken out and visually observed for turbidity every morning and then manually agitated for aeration. The bottles showing turbidity were subcultured on MacConkey agar, 5% sheep blood agar. All the negative bottles were incubated for 7 days and another blind subculture was done at the end of 7 days of incubation before reporting them as negative.[3] Any growth obtained was processed and identified using Gram-staining, colony morphology, and standard biochemical tests. Antibiotic susceptibility testing was performed according to Kirby–Bauer's disc diffusion method and interpreted according to CLSI guidelines using HiMedia (Mumbai) antibiotic discs.[4] Antifungal susceptibility was performed using disc diffusion method according to the appropriate guidelines.[5] Positive growths were further critically analyzed based on criteria to be agents of bacteremia, fungemia, and contaminants.[6] Data were manually compiled and analyzed critically for the study. The following strains were used as quality control strains:
-
Staphylococcus aureus (ATCC 25923)
-
Escherichia coli (ATCC 25922)
-
Pseudomonas aeruginosa (ATCC 27853).
Results
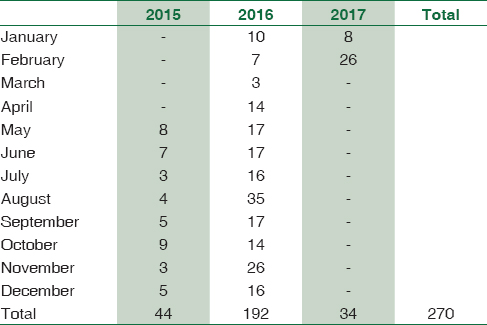
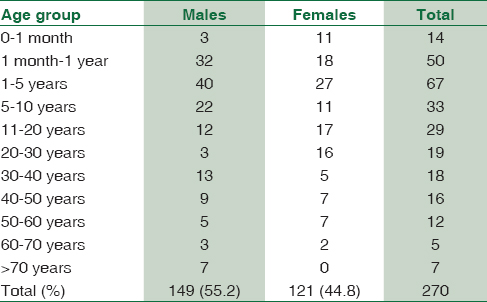
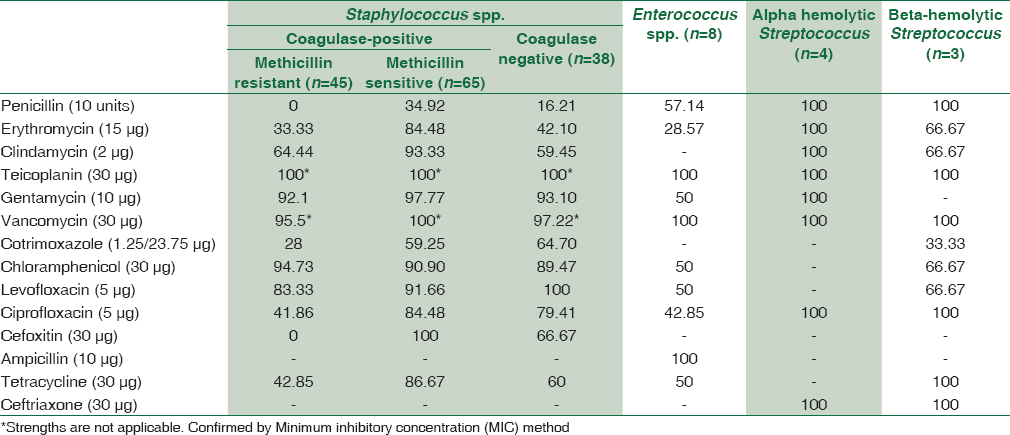
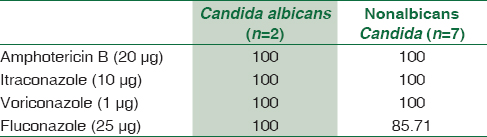
A total of 1895 consecutive samples were received from different wards and Intensive Care Units (ICUs) of the hospital. Positive aerobic bacterial growth was observed in 270 (14.24%) isolates. Thirty-one isolates (1.63%) were contaminants recovered during culture of samples. The number of pathogens recovered in each of the 22-month long study period is presented in Table 1. The age and gender distributions of the population suffering from bacteremia are described in Table 2. The mean age of patients was 15.97 ± 20 years (range 0–86 years). Gender-wise ratio of 1.23:1 was observed skewed in favor of males. From 270 isolates recovered from patients, the spectrum of microbes included 163 (60.37%) Gram-positive cocci (GPC), 98 (36.29%) Gram-negative bacilli (GNB), and 9 (3.33%) isolates of Yeasts. S. aureus was the predominant pathogen isolated from bacteremia cases followed by coagulase-negative Staphylococcus (CoNS) and Acinetobacter spp. Antibiotic susceptibility patterns for GPCs and GNBs were interpreted according to prevalent CLSI guidelines,[3] and are represented in Tables 3 and 4, respectively. Similar antifungal susceptibility profile is represented in Table 5.

Discussion
BSI is a challenging problem, and sometimes, it may be life-threatening; therefore, timely detection, identification, and antimicrobial susceptibility testing of blood-borne pathogens are one of the most important functions of diagnostic microbiology laboratory.[1] There exists a strong relationship between delay in effective initiation of therapy and in-hospital mortality of septic shock. Each hour of delay in therapy initiation is associated with an average decrease in survival of 8%.[7] This study is an attempt to analyze the bacterial profile of blood culture isolates, assess antimicrobial trends, correlate bacteremic source, and their impact which may help us devise the best ways of managing BSIs.
In the present study, the culture positivity was 14.24%. This rate of isolation is consistent with many studies from India[891011] and abroad.[1213] High culture positivity ranging from 33.9% to 52.10% were reported by various other authors.[1415] Such variation in blood culture positivity can be explained by various factors such as volume or the number of blood culture samples taken for study as explained by Lee et al.[16] All the cultures in our study yielded monomicrobial growth.
Although the mean age of patients was 15.97 years, bacteremia cases were commonly isolated from 1 to 5 year age group (24.81%) followed by those < 1 year (23.70%). Even though cumulatively males outnumber female bacteremia patients across all age groups, in our study, there were more frequent bacteremia cases noted from female neonates rather than males. Same was true for the 21–30 age group. This may be explained as men are involved in more physical activities for livelihood which predisposes them for BSI as well as they are more privileged to visit physician chamber for treatment in the Indian society.
A total of 31 isolates (1.63%) of contaminants were isolated in the concurrent study. The rate of contamination observed is below the target level suggested by Hall et al.[6] This correlates well with other studies by Archibald et al.[17] and Weinstein.[18]
In the present study, bacteremia due to Gram-positive pathogens was more common when compared to Gram-negative pathogens. This fact is corroborated by other studies as well.[192021] More than half (54.81%) of all sepsis cases were caused by Staphylococcus spp. S. aureus was the predominant pathogen (40.7%) recovered. Pre-eminence of S. aureus as a blood stream pathogen has been documented by numerous similar studies.[91921] Among these isolates, majority (~59%) were methicillin-sensitive S. aureus (MSSA) and rest 41% were methicillin-resistant S. aureus (MRSA). These figures are hardly comforting because rise in the incidence of MRSA sepsis cases spells doom for effective management of BSIs. These organisms are notorious since they do not respond to the broad class of beta-lactam antibiotics and acquire resistance to newer antibiotics quite rapidly. This effectively complicates the management of such BSIs. Over the past two decades, CoNS, the usual skin commensals are increasingly being considered bloodstream pathogens in select settings. Improper methods of blood collection and the presence of long-standing intravascular catheters are recognized as possible modes of spread of BSI by CoNS. In fact, two studies[822] reported CoNS as the most common isolate causing BSIs in ICU patients. CoNS, was the second most common isolate causing sepsis in our study as well.
Among Gram-negative pathogens, Enterobacteriaceae as a group accounted for maximum sepsis cases (41/98) with a predominance of Klebsiella species and E. coli. However, as an individual bacterial genus, nonfermenting bacilli Acinetobacter spp. (n = 30) was more predominant. Rather surprisingly, all nonfermenting bacteria combined together accounted for 58.16% of all Gram-negative sepsis. The finding is of significant concern as in the hospital settings, these isolates are associated with a high degree of antimicrobial resistance.[2324]
Among few cases of pyrexia of unknown origin investigated for blood culture, on three occasions Burkholderia pseudomallei and on two instances Salmonella typhi were recovered. Of the melioidosis cases, two were complaining of osteomyelitis and fever and one with multiple discharging sinuses. Typhoid cases were incidental findings in patients investigated for continuous low-grade fever for >7 days.
Fungemia has been documented in nine (3.33%) cases. All of them were due to Candida spp. Two were identified as Candida albicans and seven were nonalbicans Candida. The results were consistent with the study by Karunakaran et al.[25] who reported positivity rate of 3.8% for Candida spp. with a predominance of nonalbicans Candida spp. and with Kohli-Kochhar et al.[26] who reported a rate of 3% for yeast isolates. All fungemia patients were critical and from different ICUs. Risk factors for fungemia include prolonged hospital stay, hyper alimentation, hemodynamic monitoring devices, previous broad-spectrum antimicrobial therapy, and ulcerations in gastrointestinal mucosa.
An evaluation of the in vitro sensitivity pattern of GPCs was attempted. Within Staphylococcus spp., MRSA were most responsive (100%) to the action of teicoplanin, vancomycin, and chloramphenicol. Gentamicin and levofloxacin were other alternatives. MSSA isolates were highly responsive (>90%) to gentamicin, clindamycin, quinolones, and chloramphenicol besides erythromycin and tetracycline. CoNS infections were amenable to levofloxacin, gentamicin, and chloramphenicol (~90% sensitivity). Enterococcus spp. had mixed sensitivities toward gentamicin, chloramphenicol, and tetracycline. Ampicillin was however totally effective for Enterococcus spp. Alpha and beta-hemolytic Streptococcus spp. were uniformly sensitive to penicillin and other beta-lactam antibiotics.
A similar review of GNBs reveals higher susceptibility for aminoglycosides and carbapenems overall. Cephalosporins were useful for certain bacteria only. For treating BSIs caused by Enterobacteriaceae isolates, imipenem, meropenem, amikacin, and gentamicin had ~ 90%–100% sensitivity. Ciprofloxacin and cephalosporins such as cefepime and ceftriaxone were also very good alternatives to treat such cases except those perpetuated by E. coli strains. Treatment of sepsis caused by nonfermenting GNB was more successful when attempted with carbapenems, aminoglycosides, ciprofloxacin, and piperacillin-tazobactam. Pseudomonas spp. isolates were highly responsive toward ceftazidime and cefoperazone. Melioidosis cases were better managed with imipenem, ceftazidime, and chloramphenicol. Regarding fungal isolates, high sensitivity was observed for most antifungals. However, sensitivity for fluconazole was tad low with nonalbicans Candida as expected.
Effective treatment of sepsis hinges on early diagnosis and appropriate and possibly targeted antimicrobial therapy. The choice of antibiotics is based on local knowledge of bacteriological profile and antimicrobial sensitivity patterns. Beta-lactam drugs are rapidly becoming ineffective for treating BSIs due to indiscriminate and nonjudicious usage. The fact that cephalosporins are one of the most commonly used antibiotics for inpatients as well as for outpatients could be the reason for such high degree of resistance. Hence, rationalization of treatment strategies is very much warranted considering the local trends of BSIs. Poor infection control practices and inappropriate use of antibiotics are main driving forces for the spread of resistant organisms. With the shortage of newer drugs' availability and increasing resistance, use of limited option drugs such as colistin by clinicians could soon lead to the condition of so-called pan-drug-resistance.[10]
Conclusions
The present study provided much-needed information on the prevalence of bacterial pathogens in blood stream infections and their antibiotic sensitivity patterns at Port Blair in A and N islands. Specific antibiotic utilization strategies like antibiotic restriction, combination therapy, antibiotic usage according to the standard antimicrobial susceptibility testing and antibiotic recycling may help to reduce incidence of blood stream infections as well as prevent the emergence of resistance. Robust infection control practices and antibiotic stewardship programs are prime need of the hour.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Bloodstream infections and antimicrobial sensitivity patterns in a tertiary care hospital of India. Ther Adv Infect Dis. 2016;3:119-27.
- [Google Scholar]
- Bloodstream infection and endocarditis. In: Borriello SP, Murray PR, Funke G, eds. Topley and Wilson's Microbiology and Microbial Infections, Bacteriology Vol Vol. 1. (10th ed). London: Hodder Arnold ASM Press; 2005. p. :1181-235.
- [Google Scholar]
- Principles and Procedures for Blood Cultures; approved Guideline. In: CLSI Document M47-A. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement. In: CLSI Document M100-S27. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- [Google Scholar]
- Method for Antifungal disc Diffusion Susceptibility Testing of Yeast; approved Guideline. In: CLSI Document M44-A2 (2nd ed). Wayne, PA: Clinical and Laboratory Standards Institute; 2008.
- [Google Scholar]
- Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19:788-802.
- [Google Scholar]
- Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589-96.
- [Google Scholar]
- Ecology of blood stream infection and antibiotic resistance in Intensive Care Unit at a tertiary care hospital in North India. Braz J Infect Dis. 2014;18:245-51.
- [Google Scholar]
- Study of prevalence and antimicrobial susceptibility pattern in blood isolates from a tertiary care hospital in North Kerala, India. Int J Curr Microbiol Appl Sci. 2014;3:655-62.
- [Google Scholar]
- Bacteriological analysis of blood culture isolates with their antibiogram from a tertiary care hospital. Int J Pharm Sci Res. 2015;6:4847-51.
- [Google Scholar]
- Bacteriological profile and antibiogram of blood culture isolates from a tertiary care hospital of North India. Trop J Med Res. 2016;19:94-9.
- [Google Scholar]
- Bacteriologic profile and antibiogram of blood culture isolates from a children's hospital in Kabul. J Coll Physicians Surg Pak. 2014;24:396-9.
- [Google Scholar]
- Bacteriological profile of neonatal septicaemia in a tertiary hospital in Nigeria. Afr Health Sci. 2006;6:151-4.
- [Google Scholar]
- Study of prevalence and antimicrobial susceptibility pattern of bacteria isolated from blood cultures. J Biol Sci. 2009;9:249-53.
- [Google Scholar]
- Detection of bloodstream infections in adults: How many blood cultures are needed? J Clin Microbiol. 2007;45:3546-8.
- [Google Scholar]
- Blood culture contamination in Tanzania, Malawi, and the United States: A microbiological tale of three cities. J Clin Microbiol. 2006;44:4425-9.
- [Google Scholar]
- Blood culture contamination: Persisting problems and partial progress. J Clin Microbiol. 2003;41:2275-8.
- [Google Scholar]
- Bacteriologic profile and antibiogram of blood culture isolates in a pediatric care unit. J Lab Physicians. 2010;2:85-8.
- [Google Scholar]
- Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol. 2003;41:3655-60.
- [Google Scholar]
- Invasive bacteria isolates from children with severe infections in a Nigerian hospital. J Infect Dev Ctries. 2009;3:429-36.
- [Google Scholar]
- Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann Clin Microbiol Antimicrob. 2004;3:7.
- [Google Scholar]
- A retrospective study on blood stream infections and antibiotic susceptibility patterns in a tertiary care teaching hospital. Int J Pharm Pharm Sci. 2012;4:543-8.
- [Google Scholar]
- Bacteriological profile and antimicrobial susceptibility pattern of blood isolates from a tertiary care hospital in North India. Int J Pharm Res Bio Sci. 2013;2:24-35.
- [Google Scholar]
- Etiology of blood culture isolates among patients in a multidisciplinary teaching hospital in Kuala Lumpur. J Microbiol Immunol Infect. 2007;40:432-7.
- [Google Scholar]
- A ten-year review of neonatal bloodstream infections in a tertiary private hospital in Kenya. J Infect Dev Ctries. 2011;5:799-803.
- [Google Scholar]





