Translate this page into:
Carba NP as a simpler, rapid, cost-effective, and a more sensitive alternative to other phenotypic tests for detection of carbapenem resistance in routine diagnostic laboratories
Address for correspondence: Dr. Gita Nataraj, Department of Microbiology, 5th floor, MSB, KEM Hospital, Acharya Dhonde Marg, Parel, Mumbai, Maharashtra, India. E-mail: gitanataraj@gmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
PURPOSE:
Resistance to carbapenems due to carbapenemases has been increasingly noticed in Enterobacteriaceae. Clinical and Laboratory Standards Institute (CLSI) has recommended the latest Carba NP (CNP) test as a confirmatory test for carbapenemase production in Enterobacteriaceae. Low sensitivity of disk diffusion (DD) and modified Hodge test (MHT) may result in missing out of resistant strains which can adversely affect clinical management. The present study compares three phenotypic tests - CNP test, DD, and MHT for detection of carbapenemase production.
MATERIALS AND METHODS:
Four hundred consecutive, nonduplicate Enterobacteriaceae isolates were tested for carbapenem resistance using ertapenem disc (10 μg) by Kirby–Bauer DD method, MHT, and CNP. These tests were performed and interpreted as per the CLSI standards. CNP was considered to be the reference test for comparison. Sensitivity, specificity, and accuracy rates for ertapenem DD and MHT were calculated.
RESULTS:
One hundred and six out of 400 strains were positive by CNP test. Of the 294 CNP-negative strains, 28 were resistant by DD and 18 were resistant by MHT. Of the 106 CNP-positive strains, 82 were resistant and 16 were intermediate by DD while 76 were positive by MHT ertapenem DD had a sensitivity and specificity of 66.04% and 90.48%, respectively. Sensitivity and specificity of MHT were 54.72% and 93.88%, respectively. There was considerable discordance between all the three tests.
CONCLUSION:
As a rapid, simple, and cost-effective test with a greater capability greater to detect carbapenemase producers, CNP can be implemented in routine diagnostic laboratories, thereby benefiting patient care and antimicrobial stewardship.
Keywords
CarbaNP
carbapenemase
Hodge
rapid
resistance
Introduction
Carbapenemases by definition are enzymes that have the ability to hydrolyze all β-lactams including carbapenems, cephalosporins, penicillins, and sometimes aztreonam.[1] Resistance to carbapenems among Enterobacteriaceae has been attributed primarily to the production of carbapenemases and to some extent to decreased bacterial outer membrane permeability and efflux pump mechanisms.[2] Clinically they pose a problem during treatment and when linked to other resistance factors carried on the same plasmid,[1] leave almost no choice for therapy.
Carbapenemases have been reported as early as 1982 in Serratia marcescens,[3] but of late are being increasingly reported in the more common Enterobacteriaceae[1] such as Escherichia coli and Klebsiella pneumoniae. Three factors with respect to carbapenem resistance in Enterobacteriaceae are of significance:[3]
-
Enterobacteriaceae are a part of the normal flora of the gut
-
They cause a majority of infections due to Gram-negative Bacilli both in community, as well as hospital settings and
-
Carbapenem resistance in Enterobacteriaceae is carried on mobile genetic elements. Hence, the detection of carbapenemases in these strains has become an issue that we no longer can afford to ignore.
The evolution of and a rise in the prevalence of carbapenem-resistant strains may be attributed to the emergence and rapid dissemination of extended spectrum β-lactamases (ESBL)-producing strains. Because ESBL producers also demonstrated resistance to quinolones and aminoglycosides, carbapenems became the drug of choice for therapy, especially for nosocomial infections and critically ill patients.[4] The selection pressure due to the increasing use of carbapenems may be driving the rise in carbapenem resistance. Recent reports indicate the spread of these resistant strains to the community setting as well.[56]
Different phenotypic and genotypic methods have been proposed for the accurate detection of carbapenemases. The latest standard from the Clinical and Laboratory Standards Institute (CLSI) recommends Carba NP (CNP) test as the confirmatory test for carbapenemase production in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter species.[7] According to Nordmann et al.,[8] CNP test is rapid (results within 2 h) with a sensitivity and specificity of 100% when compared with polymerase chain reaction (PCR), economical, and user friendly with good reproducibility and can detect all carbapenemases equivocally.
Hence, the present study evaluates CNP test as an alternative to modified Hodge test (MHT) and ertapenem disk diffusion (DD) for the detection of carbapenemase production in Enterobacteriaceae.
Materials and Methods
Four hundred consecutive, nonduplicate Enterobacteriaceae isolates (E. coli 244; K. pneumoniae 135; Proteus spp. 14, others 7) were included in this prospective study after obtaining permission from the Institutional Ethics Committee (approval no. EC/OA-90/2013).
Identification of the strains was done using standard phenotypic criteria such as Gram's stain characteristics, growth characteristics on solid media, and results of biochemical tests.[9]
Resistance to carbapenems was detected using ertapenem (10 µg) disks by Kirby–Bauer DD method, MHT, and CNP tests. The tests were performed and interpreted as per the CLSI standards.[7]
The CNP test was performed as described by the CLSI.[7] The chemicals, antibiotic disks, and media required for performing the tests were procured from HiMedia Laboratories, Mumbai, India. The imipenem powder for CNP test was procured from Ranbaxy Laboratories, Mumbai, India.
Carba NP test
Preparation of CNP Solution A:[78] 2 ml of phenol red 0.5% w/v (0.5 g in 100 ml) was mixed with 16.6 ml clinical laboratory reagent water, vortexed and adjusted to a final pH 7.8 by adding NaOH. Phenol red is the pH indicator. A carbapenemase-producing strain breaks down imipenem into acidic products, which turns the color of the phenol red indicator to yellow. A volume of 180 µl of 10 mM ZnSO4 was added to it to obtain a final concentration of 0.1 mM ZnSO4. ZnSO4 is added to enhance the activity of metallo-beta-lactamase (MBL) carbapenemases. This increases the sensitivity of CNP to detect MBL carbapenemases.
Preparation of CNP Solution B[78] (Solution A + 6 mg/ml imipenem): Required volume of Solution B was determined (100 µl per test). The same volume of Solution A was taken, required amount of imipenem powder was weighed out and dissolved in it to yield a final concentration of 6 mg/ml imipenem.
Test Procedure:[78] 100 µl of 20 mM Tris-HCl lysis buffer was added in each of two 1.5 ml Eppendorf tubes. A volume of 10 µl of bacterial isolate was resuspended in each tube and vortexed for 1 min. The tubes are labeled Tube A and Tube B. A volume of 100 µl of solution A was added to Tube A and 100 µl of solution B was added to Tube B.
Initially, both the tubes showed red or red-orange color. Both the tubes were then incubated at 37°C for 2 h and observed for color change.
Interpretation
After 2 h, both the tubes remain red or red-orange - CNP negative Tube A remains red or red-orange, while Tube B turns light orange, yellow or dark yellow - CNP positive [Figure 1]. Both the tubes turn yellow - invalid result: A CNP-positive result implies that the tested isolate is a carbapenemase producer, while a CNP-negative result indicates that the tested isolate does not produce carbapenemases. Repeat testing is advocated in the case of an invalid result.[78]
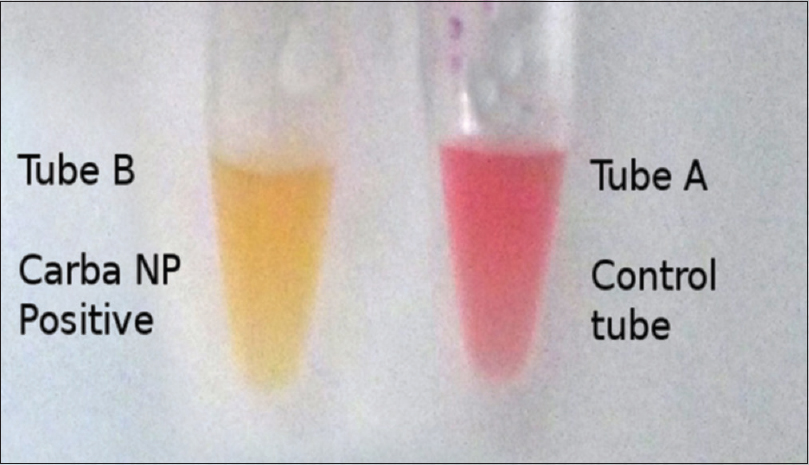
- Positive CarbaNP test result by a carbapenemase producer Tube A, the control tube, which contains Solution A shows no color change. Tube B, which contains Solution B, shows color change to yellow implying a positive CarbaNP test result
Quality control
With every panel of test isolates, quality control strains were tested too. Klebsiella pneumoniae ATCC BAA-1705 and E. coli ATCC 25922 were used as positive and negative controls, respectively. The observation and interpretation of the results were made by two independent observers to avoid observer bias. The results of all three tests were compared.
Statistical analysis
CNP was considered to be the reference test for comparison. Sensitivity, specificity, accuracy, negative, and positive predictive values for ertapenem disk diffusion and MHT were calculated.
Results
Results of the three tests performed on 400 isolates are shown [Table 1]. Two hundred and ninety-four strains were CNP susceptible, but 28 of these were resistant by DD while MHT showed 18 as resistant. While CNP detected 106 strains as carbapenemase producers, ertapenem DD detected only 82 of these as resistant and 16 as intermediate, while MHT detected only 76 as carbapenemase producers.
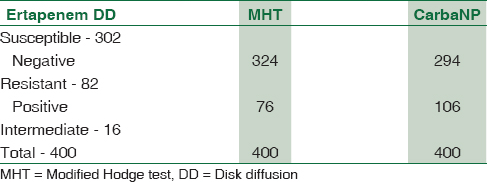
The sensitivity, specificity, PPV, and NPV of ertapenem DD and MHT were 66.04%, 90.48%, 71.43%, and 88.08% and 54.72%, 93.88%, 76.32%, and 85.19%, respectively [Tables 2 and 3].
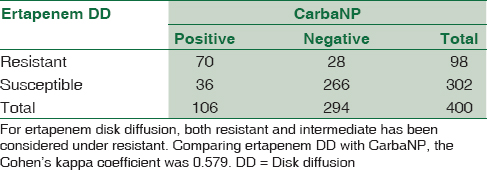
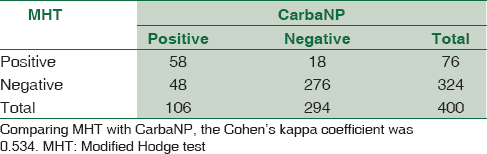
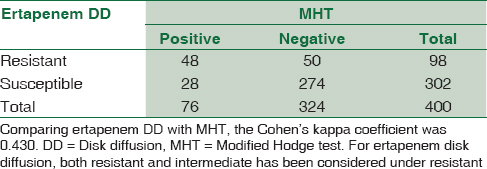
The Cohen's kappa coefficient was calculated (using GraphPad) for each pair of tests to determine the concordance of results. Comparing ertapenem DD with CNP, the kappa coefficient was 0.579 [Table 2]. Comparing MHT with CNP, the kappa coefficient was 0.534 [Table 3]. Comparing ertapenem DD with MHT, the kappa coefficient was 0.430 [Table 4].
Discussion
With phenotypic modalities such as DD and MIC determination being the most widely used techniques, in routine diagnostic laboratories to predict susceptibility or resistance of a bug to a drug, the present study compared the newly recommended CNP test to the conventional ertapenem DD and MHT to detect carbapenem resistance (due to carbapenemases) in Enterobacteriaceae. The results indicate that CNP test is rapid and more sensitive in detecting carbapenemases and at the same time cheaper than other widely used phenotypic tests.
Molecular assays such as real-time PCR which are reliable for the detection of the most prevalent carbapenemase genes, however, cannot detect resistance to carbapenems due to other as yet undetected mechanisms. They are expensive and labor intensive, making them unsuitable for regular use in routine clinical microbiology laboratories.[1011] CNP on the other hand with its reported accuracy, ease of use and rapid turnaround time (~2 h) appears to be a game changer for detecting carbapenemase mediated resistance.
Ertapenem is preferred over imipenem or meropenem for in vitro susceptibility testing due to its superior sensitivity (97% vs. 42% and 71%)[12] and has been reported to detect most of the carbapenemase producers.[1314] However, ertapenem DD suffers from low specificity (87%).[12] In the present study, ertapenem DD had a sensitivity and specificity of 66.04% and 90.48%, respectively.
MHT is used extensively as it is inexpensive and easy to perform in routine clinical microbiology laboratories. It suffers from low specificity and long turnaround time (24–48 h)[1115] and also fails to detect carbapenemase production in some isolates.[11] False-positive results can occur in isolates that produce ESBL or AmpC enzymes coupled with porin loss.[711] False-negative results are occasionally noted (e.g., some isolates producing NDM carbapenemase).[711] In the present study, sensitivity and specificity of MHT were 54.72% and 93.88%, respectively. The test is also laborious to perform and subject to inter-observer variation. The sensitivity and specificity of MHT in different studies varies.[1617]
Nordmann et al.[8] who developed the CNP test have demonstrated an excellent sensitivity and specificity when compared with the presence of resistance determining genes. On the other hand, Tijet et al.[18] in 2013 have reported an excellent specificity (100%) but a lower sensitivity (72.5%). Vasoo et al.[19] in their study have reported that the sensitivities of CNP and MHT were comparable (CNP-100% vs. MHT-98%), but CNP was more specific (CNP-100% vs. MHT-80%) and faster.
In the present study, DD when compared with CNP test yielded a kappa coefficient of 0.579, which suggests considerable discordance. Of particular importance are the results of 36 strains of Enterobacteriaceae [Table 2] which have tested positive for CNP but are susceptible by DD. Considering the 100% specificity and negative predictive value of CNP,[81819] these 36 false susceptible strains are presumably carbapenem resistant due to carbapenemase production that DD failed to detect. This can have adverse consequences in patient management. Twenty-eight strains which were resistant by DD tested negative for CNP [Table 2]. This may be due to true negativity as a consequence of mechanism of carbapenem resistance other than carbapenemases or false negative as suggested by Tijet et al.[18]
A similar discordance was obtained between MHT and CNP. Forty-eight strains were MHT negative and CNP positive [Table 3] suggesting the superior capability of CNP over MHT. Of these 48 strains, 26, 4, and 18 strains were susceptible, intermediate, and resistant, respectively, by ertapenem DD. Eighteen strains negative by CNP and positive by MHT [Table 3] might be a reflection of lower sensitivity and negative predictive value of CNP. All these 18 strains were susceptible by ertapenem DD.
CNP detects the significantly larger number of carbapenemase producers [Tables 2 and 3] that too with a much shorter turnaround time as compared to DD and MHT (CNP ~2 h, DD-16-18 h, MHT-16-20 h). CNP test is simple and gives a result on the same day with minimal reagents. It offers rapidity, cost effectiveness (CNP -  5.70 (€0.08)/test, MHT, and ertapenem DD each
5.70 (€0.08)/test, MHT, and ertapenem DD each  13.09 (€0.19)/test; noninclusive of cost of quality control strains; and greater capability (than other routine tests) to detect carbapenemase producers. Although CLSI mentions its use for epidemiology and infection control only, our experience suggests that it can be helpful in predicting carbapenemase production that eventually can aid in clinical management. Similar findings have been reported by Nordmann et al.[8] Since the choice of antibiotics is severely limited, carbapenemase-producing strains cause infections which are associated with significant mortality.[11] The rise in the prevalence of such strains prompts the incorporation of this simple, rapid, and cost-effective test in routine diagnostic microbiology laboratories.
13.09 (€0.19)/test; noninclusive of cost of quality control strains; and greater capability (than other routine tests) to detect carbapenemase producers. Although CLSI mentions its use for epidemiology and infection control only, our experience suggests that it can be helpful in predicting carbapenemase production that eventually can aid in clinical management. Similar findings have been reported by Nordmann et al.[8] Since the choice of antibiotics is severely limited, carbapenemase-producing strains cause infections which are associated with significant mortality.[11] The rise in the prevalence of such strains prompts the incorporation of this simple, rapid, and cost-effective test in routine diagnostic microbiology laboratories.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- The emergence of carbapenemase-producing Enterobacteriaceae. Infect Dis Spec Ed. 2012;1:9-13.
- [Google Scholar]
- Carbapenem-resistant Enterobacteriaceae: A menace to our most vulnerable patients. Cleve Clin J Med. 2013;80:225-33.
- [Google Scholar]
- Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791-8.
- [Google Scholar]
- High burden of antimicrobial resistance in Asia. Int J Antimicrob Agents. 2011;37:291-5.
- [Google Scholar]
- The changing face of Klebsiella pneumoniae infections in the community. Int J Antimicrob Agents. 2007;30:385-9.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 25th Informational Supplement. CLSI Document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute; 2015.
- Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18:1503-7.
- [Google Scholar]
- Koneman's Color Atlas and Textbook of Diagnostic Microbiology (6th ed). United States of America: Lippincott Williams & Wilkins; 2006. p. :213-93.
- Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother. 2008;61:548-53.
- [Google Scholar]
- Detection of carbapenemases in Enterobacteriaceae: A challenge for diagnostic microbiological laboratories. Clin Microbiol Infect. 2014;20:839-53.
- [Google Scholar]
- Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007;45:2723-5.
- [Google Scholar]
- Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2013;68:487-9.
- [Google Scholar]
- European Network on Carbapenemases. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18:432-8.
- [Google Scholar]
- Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50:477-9.
- [Google Scholar]
- The modified Hodge test is a useful tool for ruling out Klebsiella pneumoniae carbapenemase. Clinics (Sao Paulo). 2012;67:1427-31.
- [Google Scholar]
- Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50:3877-80.
- [Google Scholar]
- Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4578-80.
- [Google Scholar]
- Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol. 2013;51:3097-101.
- [Google Scholar]





