Translate this page into:
Characterization and antimicrobial susceptibility of coagulase-negative staphylococci isolated from clinical samples
Address for correspondence: Dr. Priya Datta, Department of Microbiology, Government Medical College and Hospital, Sector 32, Chandigarh - 160 030, India. E-mail: drpriyadatta@hotmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
PURPOSE:
This study has been done to speciate coagulase-negative staphylococci (CoNS) and also study their antibiotic susceptibility pattern isolated from clinical samples.
MATERIALS AND METHODS:
A total of 120 consecutive CoNS were isolated from various clinical samples such as blood, pus, wound swab, drain fluid, tracheal aspirate, peritoneal fluid, and pleural fluid over a period of 6 months. CoNS were identified by characteristic growth on media such as Blood agar and MacConkey agar. Speciation and identification were done by a range of biochemical testing such as PYR broth hydrolysis, novobiocin resistance, polymyxin B sensitivity, and then by matrix-assisted laser desorption ionization-time of flight. Antibiotic susceptibility of the isolates was done by Kirby-Bauer disk diffusion method as per CLSI 2017 guidelines.
RESULTS:
Among the 120 isolates, the most common species was Staphylococcus epidermidis (56.67%) followed by Staphylococcus haemolyticus (21.67%), Staphylococcus lugdunensis (11.67%), Staphylococcus caprae (5%), Staphylococcus cohnii (3.33%), and finally Staphylococcus vitulinus (1.67%). Good in vitro susceptibility was noted toward linezolid (100%), vancomycin (100%), teicoplanin (100%), and doxycycline (80.2%). The antibiotics to which resistance was seen were penicillin (96.5%), ciprofloxacin (57.1%), and oxacillin (45.5%). MR CoNS in our study ranged from 50% to 68.67%.
CONCLUSION:
Antibiotic resistance in CoNS is increasing toward penicillin, ciprofloxacin, and oxacillin as found in our study. The antibiotics such as vancomycin, teicoplanin, linezolid, and doxycycline which showed good in vitro susceptibility, therefore, should be kept as reserve drugs and used judiciously.
Keywords
Antibiotic susceptibility
coagulase negative staphylococci
iMLSB
Introduction
Coagulase-negative staphylococci (CoNS) have been recognized as an important agent of human infection since the past five decades.[1] Currently, there are 38 species of CoNS isolated from various human infections.[2] The important among them are Staphylococcus saprophyticus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus lugdunensis, Staphylococcus hominis, Staphylococcus capitis, Staphylococcus warneri, and Staphylococcus xylosus.[3] Earlier CoNS were taken as insignificant contaminant; now, they are regarded as major cause of nosocomial bloodstream infections, urinary tract infections, skin and soft-tissue infections, and various indwelling device-related and prosthetic implants infections.[4] The significant change in the patients’ profile, that is, increased number of premature newborns, elderly patients, chronically ill patients, and immunocompromised patients along with greater use of indwelling or implanted foreign body has made CoNS a predominant nosocomial pathogen. Colonization of skin and mucous membrane of the inpatient by multidrug resistant CoNS strain and its transmission by hands of health-care workers is critical step in the making CoNS a successful nosocomial pathogen.[5]
In addition, there is increasing antimicrobial drug resistance in CoNS limiting our therapeutic options. Human and animal origin CoNS harbor large reservoir of mobile genetic elements, which led to resistance to β-lactams, aminoglycosides, quinolones, macrolides, and tetracyclines. Resistance to β-lactams, that is, MRCoNS (methicillin-resistant CoNS) are determined by the presence of mecA gene carried on a mobile genetic element, that is, staphylococcal chromosomal cassette mec.[4]
The macrolides, lincosamides, and streptogramin B (MLSB) group of antimicrobial agents are frequently used as a drug of choice in the treatment of skin and soft-tissue infection due to CoNS and as a substitute in penicillin allergic patients. The most common mechanism of resistance to MLSB is due to erm gene which causes methylation of 23S rRNA leading to reduced binding of MLSB agents to ribosomes.[6] This erm gene encodes enzyme methylase either constitutively (cMLSB) or is inducible by certain substance (iMLSB).[7] These MLSBi strains show in vitro resistance to erythromycin while appearing susceptible to clindamycin, leading to failure to identify the resistance and clinical failure to clindamycin. Since erythromycin is an effective inducer of iMLSB resistance, therefore, using erythromycin disc in close proximity to clindamycin disc (D-test) helps in detecting this type of resistance in CoNS. Failure to identify iMLSB resistance may lead to clinical failure of clindamycin therapy. On the contrary, categorizing all erythromycin-resistant CoNS as clindamycin resistant will prevent the use of clindamycin in infections caused by truly clindamycin sensitive isolates.[5]
There are very few studies demonstrating the epidemiology and resistance profile of CoNS.[8] Thus, the aim of our study is to identify the various species of CoNS from clinical samples of blood, pus, sterile body fluids, and wound swab study their antibiotic susceptibility pattern and detect iMLSB resistance using D-test for the isolated organisms.
Materials and Methods
Study design and setting
The present study was conducted in the Department of Microbiology, Government Medical College and Hospital, Chandigarh, for 6 months from January 15, 2017, to July 15, 2017. A total of 120 consecutive, clinically significant strains of CoNS isolated in the Department of Microbiology, Government Medical College Hospital, Chandigarh, from various clinical samples were evaluated. These samples were blood wound swab, drain fluid, tracheal aspirate, peritoneal fluid, pleural fluid, and high vaginal swab.
Inclusion criteria
CoNS was considered the significant cause of bacteremia when
-
The same CoNS strain was isolated two or more times from blood culture sample within 5 days[5]
-
In case only one blood culture sample yielded growth of CoNS then at least two clinical parameter was taken into consideration – Body temperature >38°C or <36°C, TLC >12,000/μL or <2000/μL, or systolic blood pressure <90 mmHg.
These findings were taken by clinical evaluation of the patient if only one blood culture bottle grew CoNS isolate. In case of sites such as drain fluid, tracheal aspirate, and high vaginal swabs a repeat culture showing the same growth was taken as significant. Therefore, only clinical significant isolates were processed.
Strain identification
CoNS was identified by characteristic growth on blood agar, MacConkey agar, Gram staining, and various biochemical tests such as – free and bound coagulase. The following biochemical tests were performed for CoNS-PYR broth hydrolysis, novobiocin resistance, polymyxin B sensitivity, ornithine decarboxylase, and Voges Proskauer reactions.[23] The isolates were then confirmed using matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Germany).
Antibiotics susceptibility
Antibiotic susceptibility of the isolates is done by Kirby-Bauer's disc diffusion method following CLSI 2017 guidelines. The following antibiotics were used – penicillin (10 μg), oxacillin (30 μg), ciprofloxacin (5 μg), erythromycin (15 μg), clindamycin (2 μg), doxycycline (30 μg), gentamicin (10 μg), cotrimoxazole (1.25/23.75 μg), linezolid (30 μg), and teicoplanin (30 μg).[9]
In addition, the degree of resistance was defined as being high if resistance was >60%, moderate if resistance was between 60% and 30% and low resistance when the isolates showed resistance <30%.
For vancomycin-resistant CoNS, vancomycin screen agar (MHA with 6 μg/ml of vancomycin) was used.
D-TEST: All erythromycin resistant and clindamycin sensitive Staphylococcus strains were further tested by D-test. The standard recommendations for inoculum preparation and inoculation were followed on Mueller-Hinton agar. The erythromycin disc was placed at a distance of 15 mm (edge-to-edge) from clindamycin disc. After 18–24 h incubation at 37°C, appearance of clindamycin zone closest to erythromycin disc was noted.[7]
Reporting
-
Growth up to clindamycin and erythromycin discs indicates resistance to both erythromycin and clindamycin (cMLSB phenotype)
-
Demonstration of flattened CLI zone between erythromycin and clindamycin disc shows inducible clindamycin resistance, (iMLSB phenotype)
-
No flattening of clindamycin zone-negative for inducible clindamycin resistance (MS phenotype), that is, resistant to erythromycin but susceptible to clindamycin.[6]
Results
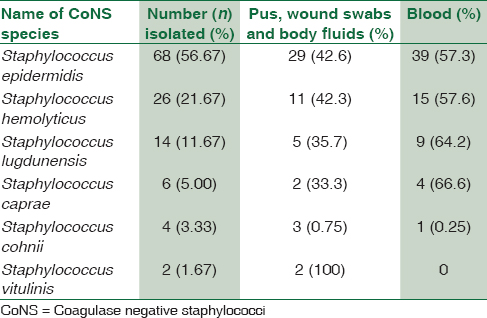
Out of the total 120 samples isolated, 68 strains of CoNS were isolated from blood, and 52 strains were from pus samples. The most common CoNS in our hospital setting was S. epidermidis (56.67%), then S. haemolyticus (21.67%) S. lugdunensis (11.67%), Staphylococcus caprae (5%), Staphylococcus cohnii (3.33%), and finally Staphylococcus vitulinus (1.67%) [Table 1]. There was 100% correlation between strains identified by biochemical reactions and MALDI-TOF MS
S. epidermidis showed high degree of resistance to penicillin (97%), oxacillin (64.7%), moderate resistance toward ciprofloxacin (58.8%), cotrimoxazole (47%), and low degree of resistance to doxycycline (23.5%), gentamicin (26.5%), and no resistance to linezolid, teicoplanin, and vancomycin [Table 2].
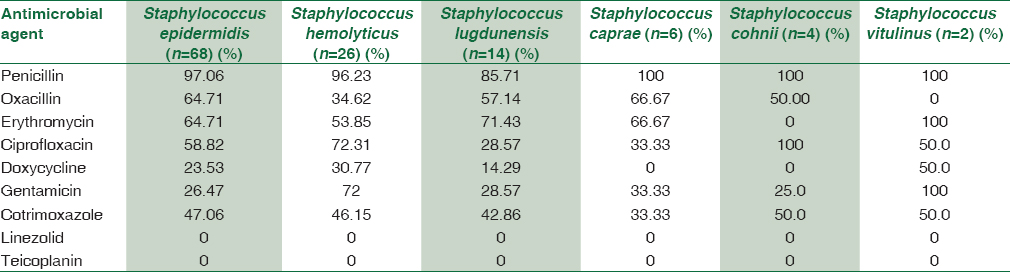
Similarly, S. haemolyticus showed a high degree of resistance toward penicillin (96.2%), ciprofloxacin (72.3%), gentamicin (72%), moderate degree of resistance toward cotrimoxazole (46.2%), oxacillin (34.6%), and doxycycline (30.7%). No resistance to linezolid, teicoplanin, and vancomycin [Table 2].
S. lugdunensis, S. caprae, and S. cohnii showed a high level of resistance toward penicillin (85%–100%), ciprofloxacin (50%–100%), and oxacillin (50%–66%). A lower degree of resistance was seen to gentamicin (25%–33%), cotrimoxazole (33%–50%), doxycycline (50%–14%), and no resistance to linezolid, teicoplanin, and vancomycin. Both the strains of S. vitulinus showed 100% resistance to penicillin, erythromycin, and gentamicin.
On PubMed search for Indian study for antibiotics resistance pattern of different CONS species in the last 10 years.
Among the 120 CoNS isolates, 43 isolates (35.8%) showed iMLS phenotype, 25 isolates (20.8%) showed cMLS phenotype, 16 isolates (13.3%) showed MS phenotype, and 36 isolates (29.9%) were sensitive to both clindamycin and erythromycin.
Out of 43 isolates showing of iMLS phenotype (D-test positive), maximum number was for S. epidermidis (62.8%) and S. haemolyticus (20.9%). Four strains of S. lugdunensis, two of S. cohnii, and one strain of S. caprae were D-test positive. No S. vitulinus showed inducible clindamycin resistance.
Discussion
The main problem in reporting of CoNS isolate from any clinical sample is to rule out colonization or contamination and report it as a true pathogen. In circumstances where false-positive CoNS reports are given, it leads to unnecessary treatment and cost and further increasing antimicrobial resistance. Isolation of CoNS strain in pure culture and repeated isolation of the same strain are useful in predicting true infections.[10] In our study, only significant CoNS were included in the study.
In our study, S. epidermidis (56.6%) was the most common species followed by S. haemolyticus (21.6%) and S. lugdunensis (11.6%). Similar results have been shown by Usha MG et al., who studied the epidemiology from blood, pus, and urine isolates and showed that S. epidermidis was the most common isolate followed by S. haemolyticus and S. lugdunensis.[11] In addition, Sheikh and Asangi et al. in their respective study showed that S. epidermidis and S. haemolyticus are the two most common isolates from the clinical samples from their respective hospital setting.[1213] In contrast, another study from northern India by Singh et al. showed S. haemolyticus (47.8%) as the most common isolate followed by S. epidermidis (33.4%).[4] This reveals that there exist variation in different species geographically. This may be due to variability in colonization by CoNS of skin and mucous membrane in diverse hospital setting owing to differences in antimicrobial pressure.
S. epidermidis and S. haemolyticus are frequent colonize moist body surfaces such as axillae, inguinal, and perineal area. These natural colonizers of skin have adapted expertly to become nosocomial pathogens because they exhibit of resistance to antibiotics and antiseptics as well as their capacity to produce biofilm.[5]
On PubMed search for Indian study on antibiotics resistance pattern of different CoNS species during the last 10 years, two studies were found – Asangi et al. (2011) and Singh S et al.[413] Both the authors showed similar resistance high resistance to penicillin for S. epidermidis, S. saprophyticus, and S. haemolyticus; similar to our study. In contrast, S. lugdunensis showed 100% resistance to penicillin by Asangi et al. whereas all the isolates of S. lugdunensis were sensitive to penicillin in the study by Singh et al. [Table 3].
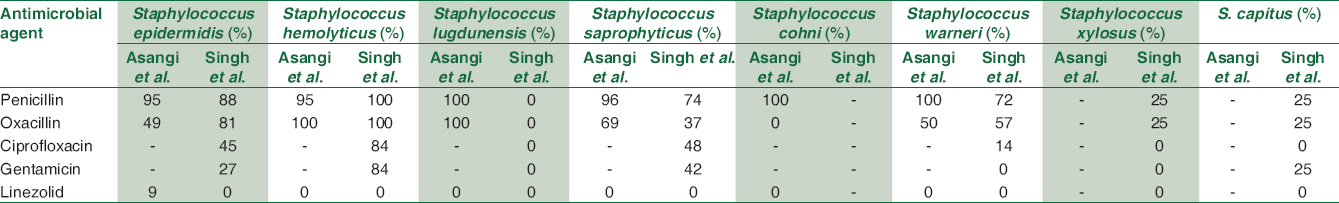
The prevalence of MRCoNS in our study ranged from 50% to 68.67%. The highest methicillin resistance was in S. caprae (66.67%), S. epidermidis (64.71%), S. lugdunensis (57.1%), and S. haemolyticus (34.6%). In the study by Asangi et al., 67.7% of CoNS were MRCoNS. This varied from S. haemolyticus and S. lugdunensis being 100% methicillin resistant to being 69.2% and S. epidermidis being 49% methicillin resistant.[13] Likewise, Singh et al. reported 100% of all S. haemolyticus strains, 81% S. epidermidis, 72% S. warneri, and 25% S. xylosus as being MRCoNS. In contrast, none of the S. lugdunensis were methicillin resistant in the study by Singh et al.[14] Similar high prevalence of MRCoNS have been reported by other authors such as Jain (48.2%) et al., Singh et al. (57.6%), and Pereira et al. (60%).[4] The high prevalence of MRCoNS not only decreases the treatment options and also assists in transfer of the resistance elements to other staphylococcal strains present in the hospital settings. The presence of methicillin resistance in CoNS has the potential or the ability to transfer resistance mechanisms to other virulent pathogens such as Staphylococcus aureus present on the skin and environment.
In addition, S. epidermidis and S. haemolyticus showed high resistance was seen to other non β-lactam antimicrobial agents – ciprofloxacin (58%–72%), gentamicin (26%–72%), and cotrimoxazole (46%–47%). Singh et al. showed similar results in their study. The resistance to ciprofloxacin varied from 84% in S. hemolyticus, 48% in S. saprophyticus, and 45% in S. epidermidis. Similar for gentamicin – 84% in S. hemolyticus, 42% in S. saprophyticus, and 27% in S. epidermidis.[14] Singh et al. showed that high resistance to ciprofloxacin, cotrimoxazole, rifampicin, and amikacin. Nearly 50% of S. lugdunensis, S. caprae, S. cohnii, and S. vitulinus isolates were MRCoNS and showed high resistance to ciprofloxacin and cotrimoxazole.[4] All CoNS isolates from our hospital setting were susceptible to linezolid, teicoplanin, and vancomycin. Linezolid was sensitive to all species of CoNS in the study by Asangi et al. and Singh et al.[1314] These are reserve drugs and should be used judicially.
Clindamycin is an excellent drug for Staphylococcal skin and soft-tissue infections and as an alternative in penicillin-allergic patient. Moreover, due to good oral bioavailability, it a preferred option for outpatient therapy and changeover after intravenous antibiotics. The differentiation of erm-mediated iMLSB phenotype isolates from isolates with MS phenotype resistance is critical issue for any clinical microbiology laboratory, because of the therapeutic implications of using clindamycin to treat a patient with an inducible clindamycin-resistant S. aureus isolate. As iMLSB phenotype is not recognized using standard susceptibility test methods; hence, D-test has to be done separately. In our institute, out the 120 CoNS isolates 20.8% showed cMLS phenotype, 35.8% showed iMLS phenotype, and 13.3% showed MS phenotype. Pal N et al. in a similar study found 46.97% of Staphylococcal isolates to be having cMLS phenotype, 23.48% were iMLS phenotype, and 29.5% showed MS phenotype.[7] Furthermore, similar findings are reported by Fiebelkorn et al. who found 30.5% of CoNS to be having inducible clindamycin resistance.[6]
Conclusion
CoNS should be regarded as pathogen of significance and not as contaminant only. Various species such as S. epidermidis, S. haemolyticus, and S. lugdunensis are causing infections these days. MR CoNS prevalence is also increasing and was in the range of 50%–68.67% for various species in this study. The other antibiotics to which resistance was noted where penicillin (96.5%), ciprofloxacin (57.15), and cotrimoxazole (44.9%). 100% susceptibility was seen towards vancomycin, teicoplanin, and linezolid. Still these drugs should be used as reserve drugs. To prevent CoNS infections, hospital infection control measures should be strengthened, and staff should be instructed to follow hand hygiene practice. Speciation of CoNS and its antibiotic susceptibility pattern will guide the clinicians in establishing their role as significant pathogens and initiate proper antimicrobial therapy based on the susceptibility pattern.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Koneman's Colour Atlas and Textbook of Diagnostic Microbiology (7th ed). Philadelphia: Wotter's Kluwer Health; 2017.
- Topley & Wilson's Microbiology & Microbial Infections Bacteriology (10th ed). London: Hodder Arnold; 2005.
- Bailey & Scott's Diagnostic Microbiology (13th ed). Missouri: Elsevier; 2014.
- Coagulase-negative staphylococci causing blood stream infection at an Indian tertiary care hospital: Prevalence, antimicrobial resistance and molecular characterisation. Indian J Med Microbiol. 2016;34:500-5.
- [Google Scholar]
- Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41:4740-4.
- [Google Scholar]
- Detection of inducible clindamycin resistance among staphylococcal isolates from different clinical specimens in Western India. J Postgrad Med. 2010;56:182-5.
- [Google Scholar]
- Changing epidemiology of coagulase-negative staphylococci in normal flora of skin. Indian J Med Microbiol. 2017;35:277-8.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. Twentieth Informational Supplement. In: CLSI Document M100. Wayne, PA: CLSI; 2017.
- [Google Scholar]
- Coagulase-negative staphylococci clinical isolates: Infectious or contaminant, that is the question. Int J Med Microbiol. 2016;1:101.
- [Google Scholar]
- Speciation of coagulase negative staphylococcal isolates from clinically significant specimens and their antibiogram. Indian J Pathol Microbiol. 2013;56:258-60.
- [Google Scholar]
- Identification and determination of coagulase negative staphylococcal species and antimicrobial susceptibility pattern of isolates from clinical specimens. Afr J Microbiol Res. 2012;6:1669-74.
- [Google Scholar]
- Nagabhushan.Speciation of clinically significant coagulase negative staphylococci and their antibiotic resistant pattern in a tertiary care hospital. Int J Biol Med Res. 2011;2:735-9.
- [Google Scholar]
- Drug sensitivity pattern of various Staphylococcus species isolated at a tertiary care hospital. Med J Armed Forces India. 2016;72:S62-6.
- [Google Scholar]





