Translate this page into:
Coagulase-Negative Staphylococci in Neonatal Blood: How Concerning?
Address for correspondence: Shampa Anupurba, MD, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, 221005, India (e-mail: shampa_anupurba@yahoo.co.in).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
Coagulase-negative staphylococci (CoNS) are being implicated as one of the leading causes of bloodstream infection (BSI). To study the spectrum, prevalence, and antimicrobial susceptibility of CoNS causing BSI in neonates.
Materials and Methods
A cross-sectional study was done in level III neonatal intensive care unit (NICU). Blood samples in automated culture bottles were processed as per the standard technique. Previously validated methods were followed for the characterization of CoNS and for AST of standard antibiotics by Kirby Bauer disk diffusion and vancomycin by agar dilution. The prevalence of causative organisms and susceptibility of CoNS were statistically analyzed. Categorical variables were compared by chi-square or Fisher's exact probability tests.
Result
In total, 1,365 blood samples (1,365 neonates) were studied, of which 383 (28.05%) were positive and 982 (71.94%) were negative. Gram-positive organisms (GPC) predominated (n = 238; 62.14%) (p < 0.001) with 41.77% (160/383) S. aureus and 13.83% (53/383) CoNS. CoNS included S. epidermidis (19, 38%), S. haemolyticus (7, 14%), S. hominis (6, 12%), S. simulans (6,12%), S. capitis (5,10%), S. cohnii (4, 8%), S. warneri (1, 2%), and S. xylosus (1, 2%). The susceptibility to netilmicin, linezolid, and vancomycin was 100% (p ≤ 0.001), and 54% (n = 27) had vancomycin MIC of 0.125 µg/mL but methicillin-resistant CoNS (MRCoNS) was 70%. Methicillin-susceptible (MS) CoNS had lower MIC of vancomycin (p < 0.05) than MRCoNS.
Conclusion
The spectrum of pathogens causing BSI in neonates is changing with predominance of GPC and among CoNS, S. epidermidis. Considerable proportion of MRCoNS with the emergence of MIC creep for vancomycin requires immediate attention.
Keywords
coagulase-negative staphylococci
bloodstream infection
NICU
neonates
Introduction
Among staphylococci, coagulase-negative staphylococci (CoNS) are frequently considered as contaminant and non-pathogenic. They are now being recognized as important human pathogens in recent decades. In the neonates admitted to the neonatal intensive care units (NICUs), CoNS are the most commonly isolated organisms.[1] There have been several reports of the isolation of this group of organisms from symptomatic severely sick patients, especially in blood cultures, thus hinting at the pathogenic potential of CoNS.[2] Additionally, a similar study in the same setup almost 15 years ago had reported CoNS as the most prevalent isolate in blood cultures although not limited to neonates.[3]
In the CoNS group, S. epidermidis is the most frequently isolated nosocomial pathogen associated with bacteremia, postoperative wounds, infection due to peripheral and central venous catheters, prosthetic valves, and other indwelling medical devices.[4,5] Various other CoNS have also been implicated in blood stream infections (BSI), including S. haemolyticus, S. hominis, S. lugdunensis, S. schleiferi, S. simulans, S. capitis, and S. warneri.[6,7] Therefore, the present study was designed to determine the spectrum of pathogens causing BSI in neonates admitted in NICU of a tertiary care center in the city of Varanasi, with special reference to proportion and antimicrobial susceptibility profile of CoNS.
Materials and Methods
Study Setting
This was a cross-sectional study conducted in the Department of Microbiology and the NICU of the Department of Pediatrics. The period of study extended from October 2017 to April 2019. The study was approved by Institutional Ethics Committee [Dean/2017/EC/193(b)]. Informed consent was taken from the parent/guardian of neonates included in the study.
Study Sample
The study included blood cultures from clinically suspected neonates with sepsis. Consecutive sampling was done. The clinical suspicion was based on neonatologist's suspicion based on the following clinical symptoms: fever or hypothermia, altered respiratory rates, feeding intolerance, and those with associated comorbidities.
Inclusion Criteria
Only inborn neonates, that is, those delivered in the same hospital and admitted in the NICU with clinical diagnosis of suspected BSI were considered for the study. Cultures showing single pathogens were considered.
Exclusion Criteria
Those not providing consent were excluded. Samples showing diphtheroids, micrococci, aerobic spore bearers, and those with multiple organisms (more than 1) were also excluded.
Isolation and Identification of Bacterial Isolates
Single blood samples from the neonates who fulfilled the inclusion criteria were aseptically collected before starting antibiotics and immediately sent to the laboratory. Culture was done in automated blood culture bottle (BACT/ALERT 3D Right Combination Module [bioMérieux, N. Carolina 27712, USA]) and incubated for 5 days. Subcultures were done from all the studied blood cultures. Those that flagged positive, subcultures were done immediately. For others, a final subculture was done on the 6th day of incubation before declaring as culture negative. The samples were subcultured on blood agar and MacConkey agar as per the standard technique.[8] No direct Gram staining was performed from the samples. The isolates were initially identified by Gram staining, catalase test, coagulase test, growth in modified Hugh and Leifson medium[8] and susceptibility to furazolidone (100 µg), resistance to bacitracin (0.04 U).[9] All the isolates, which were either slide or tube coagulase-negative were further characterized after revival from previous stock by employing two-step procedure schemes[10,11] including fermentation of mannitol, maltose, mannose, trehalose, and novobiocin (5 µg) susceptibility testing. Ornithine decarboxylase test, urease production was also performed for the identification of S. simulans, S. captis subsp. captis, and S. cohnii subsp. cohnii strains. Acetoin production (VP test) and lactose for S. haemolyticus group, growth in anaerobic condition in thioglycollate broth for S. warneri group and xylose for S. cohnii group were also performed. The scheme for speciation of CoNS is shown in ►Supplementary Table S1 (available in the online version only).[11]
Molecular Confirmation of CoNS
CoNS was confirmed by the absence of coa gene by polymerase chain reaction as shown in ►Supplementary Fig. S1 (available in the online version only) based on the method used by Goh et al.[12]
DNA Extraction
Staphylococcal DNA was isolated using the classical chloroform:phenol extraction method.[13] A few colonies from the pure culture on MHA, after 18 hours of incubation at 37°C were suspended in sterile distilled water and centrifuged. Pellets was resuspended in TE buffer followed by treatment with SDS and proteinase K, and incubation at 35°C overnight. Afterward, 5 M NaCl and CTAB were added the next day and incubated at 60°C for 10 min. Equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added and centrifuged, and the aqueous phase was collected followed by addition of an equal volume of isopropanol and kept at room temperature for 5 min. The pellets after centrifuge were washed by 70% ethanol and again centrifuged. Pellets were dried at 37°C in inverted condition and were dissolved in TE buffer and stored at −20°C.
Polymerase Chain Reaction
Sample was subjected to denaturation at 94°C for 5 minutes, 40 PCR cycles, consisting of thermal cycling for 30 seconds at 95°C, 2 minutes at 55°C, and 4 minutes at 72°C, and final extension at 72°C for 8 minutes. Bio-Rad T100 thermal cycler was used.
Image Analysis
The amplification product was separated on 1% agarose gel electrophoresis and visualized by ethidium bromide staining. Next, 1,000 bp DNA ladders were used as DNA molecular standards. Bio-Rad ChemiDoc was used.
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing (AST) of the isolates was performed against a panel of antimicrobials on Mueller-Hinton agar plates by the standard Kirby-Bauer disk-diffusion method as per Clinical and Laboratory Standards Institute (CLSI) guidelines, 2018.[14] All disks were procured from Himedia Pvt. Ltd., Mumbai, India. The antimicrobials tested were penicillin (10 units), cefoxitin (30 µg), erythromycin (15 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), ciprofloxacin (5 µg), gentamicin (30 µg), linezolid (30 µg), netilmicin (30 µg), and moxifloxacin (5 µg).
Screening for Susceptibility to Vancomycin
The susceptibility to vancomycin was screened by vancomycin screen agar containing 6 µg/mL vancomycin powder (Hi-media Pvt. Ltd., Mumbai, India). Besides, exact MIC against vancomycin susceptibility was performed by agar dilution method. The entire methodology is summarized in ►Fig. 1.
- Methodology flow diagram.
Statistical Analysis
The prevalence of causative organisms and susceptibility pattern of CoNS were statistically analyzed. Categorical variables were compared using chi-square or Fisher's exact probability tests. A value of p at 0.05 or less was considered statistically significant. SPSS Statistics Version 21 (Armonk, NY: IBM Corp.) was used for the analysis.
Results
A total of 1,365 blood samples from 1,365 neonates was studied. Of these, 982 (71.94%) including culture contaminants (54/1365; 3.95%) were reported as negative. Of the remaining 383 (28.05%) samples, 160 (41.77%) showed the presence of S. aureus and 53 (13.83%) as CoNS, which were isolated from blood culture as monomicrobial entities.
Gram-positive organisms (n = 238; 62.14%) were significantly more (p < 0.001) than gram-negative organisms (n = 118; 30.8%) and Candida spp. (n = 27; 7.05%). Gram-positive organisms included Staphylococcus aureus, CoNS, Enterococcus faecalis, Streptococcus spp., and gram-negative organisms included Klebsiella pneumoniae, Acinetobacter spp., Escherichia coli, Pseudomonas spp., and Citrobacter spp.
Of the total 53 confirmed CoNS, 3 isolates could not be retrieved from stock and the remaining 50 were biochemically characterized as S. epidermidis (19, 38%), followed by S. haemolyticus (7, 14%), S. hominis (6, 12%), S. simulans (6, 12%), S. capitis (5, 10%), S. cohnii (4, 8%), S. warneri (1, 2%), S. xylosus (1, 2%), and 1 isolate was unidentified because of aberrant characteristics (►Table 1). The clinical outcome of the neonates affected with CoNS showed 1.88% (1/53) mortality.
| Species of CoNS | Number of isolates (%) |
|---|---|
| S. epidermidis | 19 (38) |
| S. haemolyticus | 7 (14) |
| S. hominis | 6 (12) |
| S. simulans | 6 (12) |
| S. capitis | 5 (10) |
| S. warneri | 1 (2) |
| S. cohnii | 4 (8) |
| S. xylosus | 1 (2) |
| Unidentified | 1 (2) |
| Total isolates | 50 |
Note: The most common CoNS isolate was S. epidermidis, followed by S. haemolyticus, S. hominis, S. simulans, S. capitis, S. cohnii, S. warneri, and S. xylosus.
Susceptibility Pattern of CoNS
Antimicrobial susceptibility pattern of CoNS isolated from neonates showed a high susceptibility to vancomycin (100%), linezolid (100%), netilmicin (100%) as shown in ►Fig. 2. This was statistically significant (p ≤ 0.001) when compared with the susceptibility of other drugs used, that is, gentamicin (80%), moxifloxacin (72%), trimethoprim/sulfamethoxazole (64%), ciprofloxacin (60%), methicillin (30%), erythromycin (26%), and penicillin (2%).
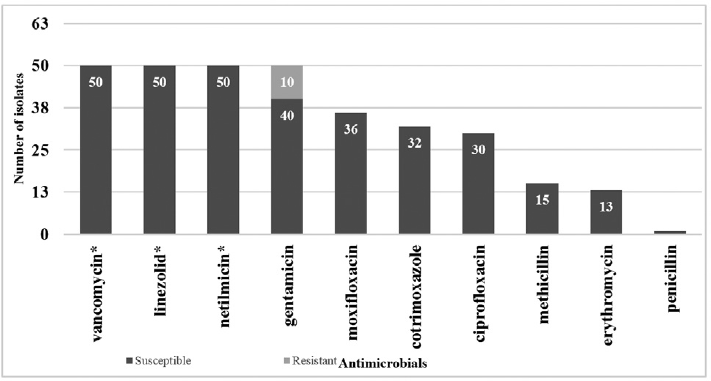
- Susceptibility pattern of all CoNS isolates. Note: Susceptibility to vancomycin (50, 100%), linezolid (50, 100%), netilmicin (50, 100%) was *statistically significant (p ≤ 0.001) when compared with susceptibility of other drugs used, i.e., gentamicin (40, 80%), moxifloxacin (36, 72%), trimethoprim/sulfamethoxazole (32, 64%), ciprofloxacin (30, 60%), methicillin (15, 30%), erythromycin (13, 26%), and penicillin (1, 2%).
Vancomycin MIC for all CoNS Isolates
In this study, 54% (n = 27) of isolates had an MIC of 0.125 μg/mL, 30% (n = 15) had an MIC of 0.25 μg/mL, and 16% (n = 8) had an MIC of 0.5 μg/mL, as shown in ►Fig. 3. In comparison to MRCoNS, MSCoNS had lower MIC of vancomycin that was significant (p < 0.05) and only MRCoNS isolates (22.85%) had an MIC of 0.5 μg/mL.
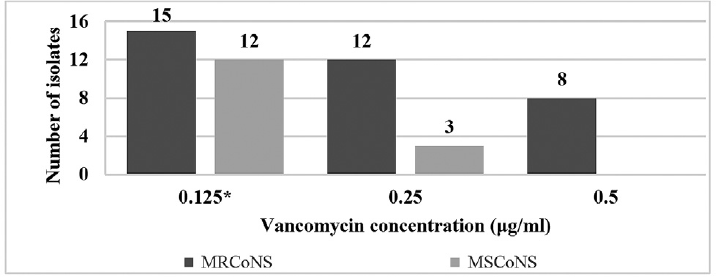
- Vancomycin MIC for all CoNS isolates. Note: 54% (n = 27) of isolates had an MIC of 0.125 μg/mL, 30% (n = 15) had an MIC of 0.25 μg/mL, and 16% (n = 8, all MRCoNS) had an MIC of 0.5 μg/mL. *In comparison to MRCoNS, MSCoNS isolates had lower MIC of vancomycin that was significant (p < 0.05).
Discussion
This study showed a proportion of 13.83% CoNS in neonatal BSI. As compared with other studies, in a study by Nazir[15] from India, out of 356 neonates screened, coagulase-negative staphylococci (30.27%) were the most common organisms isolated. In another study from India, Muthukumaran[16] screened 5,047 neonates and 38.6% of the isolates were CoNS. Thapa et al[17] showed that 56 blood samples from neonates yielded Acinetobacter species 18 (32.1%), followed by S. aureus (11, 19.6%) and CoNS (6, 11.11%). Low prevalence of CoNS may be attributed to the adherence to better infection control practices and stringent aseptic collection of blood sample, decreasing the possibility of growth of CoNS as contaminants.
In this study of 50 CoNS isolates, S. epidermidis 19 (38%) was the most common species followed by S. haemolyticus (7, 14%). In a study from India by Goyal et al,[10]S. epidermidis (41%) was the predominant isolate followed by S. haemolyticus (14.7%) and in a study by Sah et al,[11] out of 76 CoNS isolated from blood samples, S. epidermidis (42.1%) was predominant similar to our study. However, in studies by Alex et al[18] and Chaudhury et al[19] from India, S. haemolyticus was the predominant isolate. This difference in the predominant CoNS species may be due to change in geographical location and patient population.
In the International Nosocomial Infection Control Consortium (INICC),[20] of the 4.67% coagulase-negative staphylococci isolated from intensive care units, 91.4% were nonsusceptible to oxacillin. Methicillin resistance in CoNS isolated by Alex et al[18] was 73.9%. The results of this study were similar to those of aforementioned studies with 70% (n = 35) methicillin resistance and ranged from 40 to 100% for various species of CoNS isolated. In this study, although 70% isolates were MRCoNS, the outcome was good and one neonate having BSI with CoNS died. MRCoNS is important because in NICU an outbreak can occur when a methicillin-resistant strain is transmitted to other neonates and often it occurs when a healthcare worker is colonized with resistant strain causing increased morbidity. MRCoNS infection also narrows down the treatment options in neonates to vancomycin and linezolid-like reserve drugs, which may result in vancomycin-resistant strains.
In this study, vancomycin MIC ranged from 0.125 to 0.5 μg/mL and MSCoNS had lower MIC of vancomycin that was significant (p < 0.05) in comparison to MRCoNS isolates indicating a steady build-up of increased MIC to vancomycin. In a study by Paiva,[21] MICs of vancomycin for CoNS ranged from 0.25 to 2 μg/mL, while most MICs determined were 1 μg/mL or lower (128 isolates, 98.5%) by BMD, and MRCoNS had higher MIC than MSCoNS for vancomycin. In another study by Mashaly et al,[22] all CoNS isolates were susceptible to vancomycin but there was an increase in vancomycin MIC range of oxacillin resistant isolates (0.25–2 µg/mL) in comparison to oxacillin-sensitive isolates (0.25–1 µg/mL). More studies are needed to evaluate the vancomycin MIC creep in MRCoNS as seen in methicillin-resistant S. aureus.
Laboratory tests performed for coagulase-negative staphylococci neither resolves whether an isolate is a pathogen or contaminant nor supports a diagnosis of an infectious process. A study by Paolucci et al[23] showed growth of same organism when two blood samples were collected with a gap of 15 to 30 minutes, from different peripheral sites of neonates, once sepsis was suspected suggesting that a single site blood culture should be sufficient to document sepsis.
However, the study was not without limitations. We could not identify whether the CoNS isolated was true pathogen or a probable contaminant due to non-availability of paired blood samples. However, most of the neonates improved on administration of antibiotic based on the causative agent of BSI as identified in the study. Second, methicillin resistance was not identified by the presence of mecA/mecC gene and genotypic characterization based on SCCmec was not done, which could have predicted the mechanism of resistance in these isolates. Nonetheless, epidemiological study of this type are essential to know the current scenario in the local settings for improved care and outcome. Therefore, based on this study, we recommend speciation of CoNS in diagnostic laboratories and judicious use of antibiotics to curb the emergence of creep in MIC to high-end antibiotics in CoNS.
Conclusion
The study showed the predominance of gram-positive organisms (62.14%) as compared with gram-negative organisms (30.8%) in neonatal BSI. Among CoNS, S. epidermidis (38%) was a significant cause of BSI in neonates. Alarming resistance was seen with MRCoNS being isolated in 70% of the cases. Though vancomycin susceptibility was seen in all CoNS isolates, vancomycin MIC for MRCoNS was higher than MSCoNS, indicating a steady build-up of increased MIC to vancomycin. Therefore, based on this study, further research should be planned to document the emerging drug resistance and the spectrum of species under CoNS.
Ethical Approval
The study was approved by the Institutional Ethics Committee.
Conflict of Interest
None declared.
References
- Coagulase-negative staphylococcal infections in the neonatal intensive care unit. Infect Control Hosp Epidemiol. 2011;32(07):679-686.
- [CrossRef] [PubMed] [Google Scholar]
- Significance of coagulase negative Staphylococcus from blood cultures: persisting problems and partial progress in resource constrained settings. Iran J Microbiol. 2016;8(06):366-371.
- [Google Scholar]
- Bacteriological profile and antimicrobial resistance of blood culture isolates from a university hospital. JIACM. 2007;8(02):139-143.
- [Google Scholar]
- Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int J Antimicrob Agents. 2006;28(Suppl. 01):S14-S20.
- [CrossRef] [PubMed] [Google Scholar]
- Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23(01):73-98.
- [CrossRef] [PubMed] [Google Scholar]
- Phenotypic characterisation of coagulase-negative staphylococci isolated from blood cultures in newborn infants, with a special focus on Staphylococcus capitis. Acta Paediatr. 2017;106(10):1576-1582.
- [CrossRef] [PubMed] [Google Scholar]
- Speciation of coagulase negative Staphylococcal isolates from clinically significant specimens and their antibiogram. Indian J Pathol Microbiol. 2013;56(03):258-260.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of various methods for differentiation of staphylococci and micrococci. J Clin Microbiol. 1984;19(06):875-879.
- [CrossRef] [PubMed] [Google Scholar]
- Simple and economical method for speciation and resistotyping of clinically significant coagulase negative staphylococci. Indian J Med Microbiol. 2006;24(03):201-204.
- [CrossRef] [PubMed] [Google Scholar]
- Simple and economical method for identification and speciation of Staphylococcus epidermidis and other coagulase negative Staphylococci and its validation by molecular methods. J Microbiol Methods. 2018;149:106-119.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30(07):1642-1645.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular cloning–A laboratory manual. Vol 545. New York: Cold Spring Harbor Laboratory; 1982. p. :S.
- [Google Scholar]
- Performance standards for antimicrobial susceptibility testing. In: CLSI supplement M100 (28th). Wayne, PA: Clinical and laboratory standards institute; 2018.
- [Google Scholar]
- Neonatal sepsis due to coagulase negative staphylococci: a study from Kashmir valley, India. Int J Contemp Pediatrics. 2019;6:650-655.
- [CrossRef] [Google Scholar]
- Mortality profile of neonatal deaths and deaths due to neonatal sepsis in a tertiary care center in southern India: a retrospective study. Int J Contemp Pediatrics. 2018;5:1583-1587.
- [CrossRef] [Google Scholar]
- Changing trend of neonatal septicemia and antibiotic susceptibility pattern of isolates in Nepal. Int J Pediatr. 2019;2019 3784529
- [CrossRef] [PubMed] [Google Scholar]
- Speciation and antibiotic susceptibility testing of coagulase-negative staphylococci at a tertiary care teaching hospital. Int J Curr Microbiol Appl Sci. 2017;6(05):713-721.
- [CrossRef] [Google Scholar]
- In vitro activity of antimicrobial agents against oxacillin resistant staphylococci with special reference to Staphylococcus haemolyticus. Indian J Med Microbiol. 2007;25(01):50-52.
- [CrossRef] [PubMed] [Google Scholar]
- International Nosocomial Infection Control Consortium (INICC) report, data summary of 45 countries for 2013-2018, Adult and Pediatric Units, Device-associated Module. Am J Infect Control. 2021;49(10):1267-1274.
- [Google Scholar]
- Vancomycin MIC for methicillin-resistant coagulase-negative Staphylococcus isolates: evaluation of the broth microdilution and Etest methods. J Clin Microbiol. 2010;48(12):4652-4654.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin heteroresistance in coagulase negative Staphylococcus blood stream infections from patients of intensive care units in Mansoura University Hospitals, Egypt. Ann Clin Microbiol Antimicrob. 2017;16(01):63.
- [CrossRef] [PubMed] [Google Scholar]
- How can the microbiologist help in diagnosing neonatal sepsis? Int J Pediatr. 2012;2012 120139
- [CrossRef] [PubMed] [Google Scholar]






