Translate this page into:
Detection of Carbapenemase Production in Gram-negative Bacteria
Address for correspondence: Dr. Purva Mathur, E-mail: purvamathur@yahoo.co.in
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The greatest threat to antimicrobial treatment of infections caused by Gram-negative bacteria is the production of carbapenemases. Metallo-beta-lactamases and plasmid-mediated serine carbepenemases like Klebsiella pneumonia carbapenemase are threatening the utility of almost all currently available beta-lactams including carbapenems. Detection of organisms producing carbapenemases can be difficult, because their presence does not always produce a resistant phenotype on conventional disc diffusion or automated susceptibility testing methods. These enzymes are often associated with laboratory reports of false susceptibility to carbapenems which can be potentially fatal. Moreover, most laboratories do not attempt to detect carbapenemases. This may be due to the lack of availability of guidelines and procedures or lack of knowledge and expertise. Because routine susceptibility tests may be unreliable, special tests are required to detect the resistance mechanisms involved. This document describes the standard methodology for detection of various types of carbapenemases, which can be put to use by laboratories working on antimicrobial resistance in Gram-negative bacteria.
Keywords
Carbapenemases
Gram-negative bacteria
Klebsiella pneumoniae carbapenemase
metallo-beta-lactamases
New Delhi metallo-beta-lactamase
INTRODUCTION
Carbapenemases are diverse enzymes that vary in their ability to hydrolyze carbapenems and other beta-lactams. Detection of carbapenemase is a crucial infection control issue because they are often associated with extensive antibiotic resistance, treatment failures and infection-associated mortality. Among the beta-lactamases, the carbapenemases, especially transferrable metallo-beta-lactamases (MBLs) are the most feared because of their ability to hydrolyze virtually all drugs in that class, including the carbapenems. The major concern is with transmissible carbapenemases. The transmissible enzymes can be acquired unpredictably by important nosocomial pathogens such as Pseudomonas aeruginosa, Acinetobacter baumannii and members of the family Enterobacteriaceae. The chromosomal enzymes occur predictably in less common pathogens such as Stenotrophomonas maltophilia, Aeromonas spp., Chryseobacterium spp., and others.[1]
In addition to their resistance to all beta-lactams, the MBL producing strains are frequently resistant to aminoglycosides and fluoroquinolones.[2] However, they usually remain susceptible to polymyxins. Unlike carbapenem resistance due to several other mechanisms, the resistance due to MBL and other carbapenemase production has a potential for rapid dissemination, as it is often plasmid-mediated.[23] Consequently, the rapid detection of carbapenemase production is necessary to initiate effective infection control measures to prevent their dissemination.
This document details the recommended procedures for testing for the production of carbapenemases by Gram-negative bacteria. Many isolates producing carbapenemases have carbapenem minimal inhibitory concentration (MICs) around the susceptible breakpoints making resistance difficult to detect, particularly with automated systems. Therefore, disc diffusion zone breakpoints are needed as first-line screening methods. The procedures described herein can be divided into screening and confirmatory tests.
TESTS PERFORMED FOR THE DETECTION OF CARBAPENEMASES
Screening test
-
Disc diffusion method
-
E-test for MIC determination
-
Automated antimicrobial susceptibility systems: Vitek-2 (Biomerieux, France), Phoenix, MicroScan Walk-Away, Micronaut, Sensititre.
Confirmatory test
-
Modified Hodge test (MHT)
-
Ethylene diamine tetraacetic acid (EDTA) impregnated disc test
-
2-mercaptopropionic acid disc test
-
Boronic acid disc test.
Molecular methods
-
Polymerase chain reaction (PCR) (conventional-Applied Biosystems-California, United States, Gradient- GenePro, Hong Kong)
-
Sequencing.
MATERIALS AND REAGENTS REQUIRED FOR PHENOTYPIC METHODS
-
Petri-plates (Hi-Media, Mumbai/Similar standards)
-
Antibiotic discs (Becton Dickson, Franklin lakes, New Jersey, USA/Similar standards)
-
Incubator (Widsons Scientific Works, New Delhi, India)
-
E-test strips (Biomerieux, France)
-
Swab sticks
-
Normal saline (Biomerieux, France)
-
0.5 M EDTA (Fischer Scientific, Hampton, New Hamptonshire, USA, Catalogue no-12635)
-
2-mercaptopropionic acid (3 μl) (Hi-Media, Mumbai, India-RM4725-100G)
-
Micropipettes (20-200 μl, 0.5-10 μl) – (Eppendorf, Hamburg, Germany)
-
Phenyl boronic acid (Sigma Aldrich, St. Louis, MO, USA-78181-10G)
-
Dimethyl Sulfoxide (20 mg of phenyl boronic acid is dissolved in 1 ml of DMSO (Ameresco, Massachusetts, USA Lot no-0031C474)
-
0.5 McFarland standard.
PREPARATION OF MEDIA
-
MacConkey preparation (Hi-Media, Mumbai, India, catalog no-M008) - For 1 L of solution add 51.52 g of the MacConkey powder. Autoclave it for 15 min at 15 lbs pressure at 121°C. Mix it well before pouring
-
Blood agar preparation (Biomerieux, France, catalogue no-43041) - Ready to use plates may be used
-
Mueller Hinton Agar (MHA) preparation - (Hi-Media, Mumbai, India, catalog no-M173) - For 1 L of solution add 38 g of the MHA powder. Autoclave it for 15 min at 15 lbs pressure at 121°C. Mix it well before pouring.
Screening tests
In the laboratories, a carbapenem-intermediate or - resistant result should always raise the suspicion of possible carbapenemase production, as should reduce carbapenem susceptibility within the susceptible range in isolates of members of the family Enterobacteriaceae and Acinetobacter spp. The best carbapenem for screening is unknown.
The following parameters are used to suspect carbapenemase production:[45]
-
For Escherichia coli or Klebsiella sp., an imipenem MIC of ≥2 μg/ml
-
For Enterobacter sp., Serratia sp., and Citrobacter sp., an imipenem MIC of ≥4 μg/ml
-
For isolates of Acinetobacter sp., an imipenem MIC of ≥8
-
For isolates of P. aeruginosa, an imipenem MIC of ≥16 μg.
The 2013 recommendations of Clinical and Laboratory Standards Institute (CLSI) for zone diameters of ertapenem (10 μg) and meropenem (10 μg) can be followed for carbapenem resistance screening. A diameter for ertapenem of <19 mm and that for meropenem of <16 mm is indicative of carbapenemase production.[456]
PROCEDURE FOR PHENOTYPIC METHODS
Disc diffusion methods
-
Make 0.5 McFarland of standard suspension of test strain in normal saline
-
With the help of sterile swab stick, test strain is streaked as a lawn on the MHA plates
-
Place the desired antibiotic disc (imipenem, ertapenem, and meropenem) for the desired test
-
Incubate the plate at 37°C for 24 h
-
Measure the zone of inhibition around the antibiotic disc for the test strain with the help of scale
-
Interpret the results of the test strains with the CLSI guidelines.
E-test
This test is performed to detect the MIC of the test strain for a particular antimicrobial.
-
Make 0.5 McFarland of standard suspension of test strain was made in normal saline
-
With the help of sterile swab stick, test strain is streaked as a lawn on the MHA plates
-
Place the desired E-test (Biomerieux, France) for the desired test
-
Incubate the plate at 37°C for 24 h
-
Note the zone of inhibition made by for the test strain [Figure 1]
-
Interpret the results of the test strains with the CLSI guidelines.
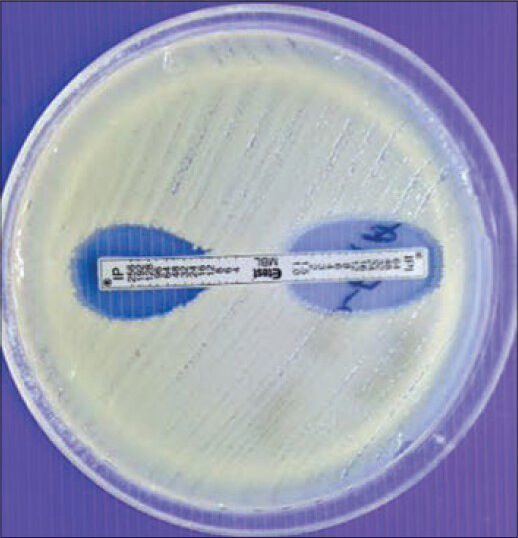
- E-test showing carbapenemase production
AUTOMATED METHODS
These are used as per manufacturer's instructions.
Phenotypic confirmation for cabapenemase production
Modified Hodge test
The MHT detects carbapenemase production in the isolates of Enterobacteriaceae.[145]
-
Make 0.5 McFarland of standard suspension of E. coli ATCC 25922 (indicator organism) in normal saline
-
Add 300 μl of 0.5 McFarland suspension of E. coli ATCC 25922 in 2700 μl sterile normal saline, to make a 1:10 dilution. With the help of sterile swab stick, this 1:10 dilution of indicator strain (E. coli ATCC 25922) is streaked as a lawn on MHA plate
-
A 10 μg meropenem/10 μg ertapenem disk is placed in the middle of the agar plate after background lawn streaking
-
With the help of sterile loop, pick three or four colonies of the test strain and streak on the plate from meropenem/imipenem disc toward the periphery
-
Positive control (Klebsiella pneumoniae ATCC 1705) and negative control (K. pneumoniae ATCC 1706) are streaked on the same plate
-
Examine the plate after 24 h of incubation
-
Carbapenemase producing isolate is detected by the MHT when the test isolate produces the enzyme and allows the growth of the carbapenem susceptible E. coli ATCC 25922 strain towards the disk.
Interpretations of the diameters of zone of inhibition are as follows
Modified Hodge test positive test
It has a clover leaf-like indentation of the E. coli 25922 growing along the test organism growth streak within the disk diffusion zone indicating that this isolate is producing a carbapenemase [Figure 2].
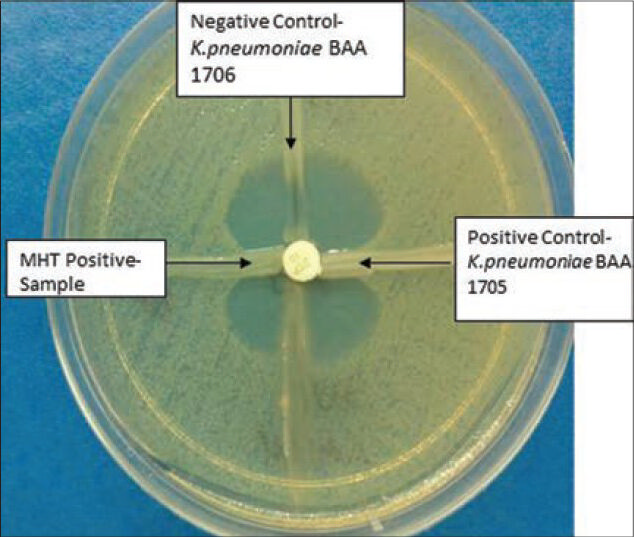
- Modified Hodge test
Modified Hodge test negative test
It has no growth of the E. coli 25922 along the test organism growth streak within the disc diffusion indicating that this isolate is not producing a carbapenemase [Figure 2].
QC testing
-
Positive control: K. pneumonia ATCC BAA 1705
-
Negative control: K. pneumonia ATCC BAA 1706.
For isolates positive with the ertapenem or meropenem disk screen and positive with the MHT, MIC test should be performed prior to reporting any carbapenem results, as recommended by CLSI. For isolates that are MHT positive but test susceptible to a carbapenem (ertapenem MIC ≤2 μg/ml; imipenem MIC ≤4 μg/ml or meropenem MIC ≤4 μg/ml), the carbapenem MIC should be noted. If the MHT is negative, the carbapenem MICs is interpreted using current CLSI interpretive criteria.
DETECTION OF METALLO-BETA-LACTAMASES
Metallo-beta-lactamase E-test
This will be performed using the following combinations of E-tests:[145]
-
Imipenem: Imipenem/EDTA
-
Meropenem: Meropenem/EDTA
-
Imipenem: Imipenem + 2 mercaptopropionic acid
-
Meropenem: Meropenem + 2 mercaptopropionic acid.
-
Make 0.5 McFarland suspension of the test strain in normal saline
-
With the help of sterile swab stick, test strain is streaked as a lawn on the MHA plates
-
Place the above E-test strips (Biomerieux, France) on the plate
-
Incubate the plate at 37°C for 24 h
-
A reduction of carbapenem MIC by ≥ 3 two-fold dilutions in the presence of EDTA or “phantom” zone between the two gradient sections or deformation of the IP ellipses is an indicator of MBL producer [Figures 1 and 3].
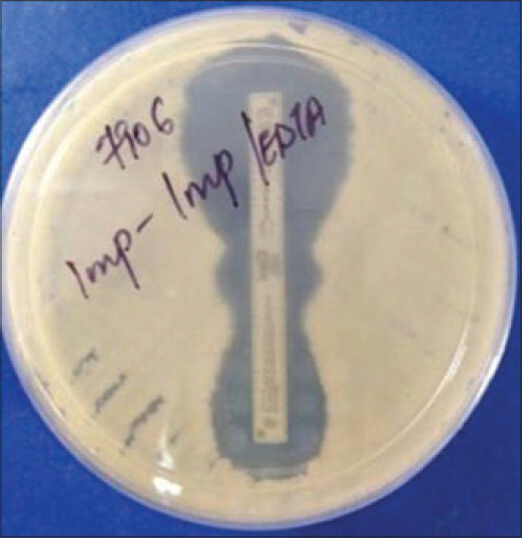
- Phantom zone in E-test indicating carbapenemase production
Zone enhancement with ethylenediaminetetraacetic acid impregnated imipenem and ceftazidime discs
Preparation of 0.5 M ethylenediaminetetraacetic acid
-
Dissolve 186.1 g of disodium EDTA. 2H2O in 1000 ml of distilled water and adjust it to pH 8.0
-
If the final solution requires any pH adjustments, add few pellets of NaOH
-
The mixture is sterilized by autoclaving.
Procedure of the test
-
Make 0.5 McFarland of standard suspension of test strain in normal saline
-
With the help of sterile swab stick, test strain is streaked as a lawn on the MHA plates
-
Place two imipenem and two ceftazidime disk on the MHA plate
-
Put 4 μl of EDTA on one of the respective disk to obtain the concentration of 750 μg
-
The inhibition zones of imipenem, ceftazidime, imipenem-EDTA and ceftazidime-EDTA discs is compared after 24 h of incubation
-
Difference in the zone diameter of ≥5 mm between imipenem/ceftazidime disc alone and imipenem-EDTA/ceftazidime-EDTA disc respectively is considered as positive [Figure 4].[145]
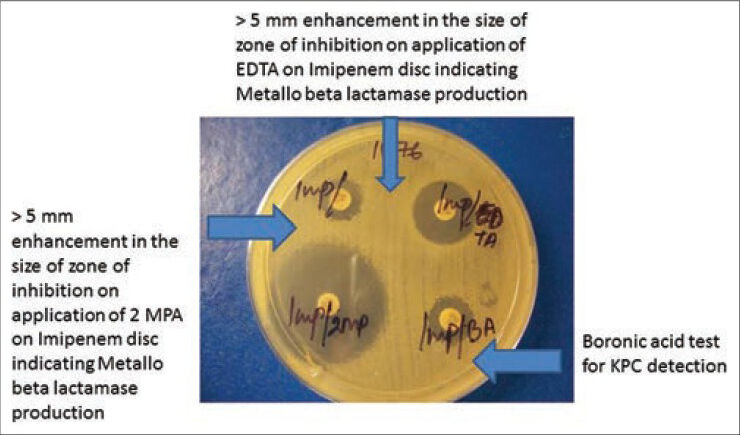
- Different methods of detection of metallo-beta-lactamase and Klebsiella pneumoniae carbapenemases
2-mercaptopropionic acid inhibition (2-MPA) test
-
Make 0.5 McFarland of standard suspension of test strain in normal saline
-
With the help of sterile swab stick, test strain is streaked as a lawn on the MHA plates
-
Two disk of ceftazidime (30 μg), two of 10 μg imipenem, two of 30 μg cefepime are placed at the distance of 50 mm
-
3 μl of 2-mercaptopropionic acid is put on one of the each disk respectively
-
Plates are incubated at 37°C for 24 h
-
The diameter of the growth-inhibitory zone around a beta-lactam disk with 2-meracptopropionic acid is compared with that around the corresponding beta-lactam disk without 2-mercaptopropionic acid
-
The test is considered positive for the detection of MBL enzyme production when the diameter of the growth-inhibitory zone around a beta-lactam disk with 2-mercaptopropionic acid is ≥5 mm larger than that around a disk containing the beta-lactam substrate alone [Figure 4].[145]
SPECIAL TESTS FOR KLEBSIELLA PNEUMONIAE CARBAPENEMASES PRODUCTION
Infections caused by bacteria-producing K. pneumoniae carbapenemases (KPCs) are becoming an increasingly significant problem worldwide since the first detection of these enzymes more than a decade ago. Although KPCs do not represent the first or the sole Mechanism of carbapenem resistance, they are remarkable because they are often not detected by routine susceptibility screening and possess an exceptional potential for dissemination.[6]
In addition to the infection control challenges that have arisen, infections caused by these organisms present clinicians with serious treatment challenges, due to limited antibiotic options.
Ertapenem has been recommended as the best screening agent for KPC detection because KPC producers are usually insusceptible to ertapenem, but may remain susceptible to other carbapenems.
BORONIC ACID TEST FOR KLEBSIELLA PNEUMONIAE CARBAPENEMASES DETECTION
Preparation of boronic acid - For stock preparation, adds 20 μg of phenyl boronic acid in 1 ml of dimethyl sulfoxide.
Working solution - 20 μl of the boronic acid is used (containing 400 μg of boronic acid).
Control strains - K. pneumonia BAA 1705 (positive control), K. pneumonia BAA 1706 (negative control).
Test procedure
-
Make 0.5 McFarland suspension of test strain in normal saline
-
With the help of sterile swab stick, test strain is streaked as a lawn on the MHA plates
-
The tests is performed by inoculating MHA by the standard diffusion method and placing disks containing eight different beta-lactams (imipenem, meropenem, ertapenem, cefepime, cefoxitin, cefotetan, cefotaxime and ceftazidime) with or without boronic acid onto the agar
-
The agar plates are incubated at 37°C overnight
-
The diameter of the growth-inhibitory zone around a beta-lactam disk with boronic acid is compared with that around the corresponding beta-lactam disk without boronic acid
-
The test is considered positive for the detection of KPC enzyme production when the diameter of the growth-inhibitory zone around a beta-lactam disk with boronic acid is ≥5 mm larger than that around a disk containing the beta-lactam substrate alone [Figure 4].
Genotypic methods
-
PCR
-
Sequencing.
POLYMERASE CHAIN REACTION - BASED IDENTIFICATION OF CARBAPENEMASE GENES
Material and reagents required for genotypic method (PCR)
-
PCR machine
-
Sterile micro-tips (0.2 ml, 1.7 ml)
-
Taq polymerase
-
Primers
-
Micropipettes (20-200 μl, 0.5-10 μl, 100-1000 μl, 10-100 μl)
-
Laminar flow chamber
-
MgCl2
-
Buffers
-
dNTPs
-
PCR grade water
-
Agarose
-
Gel electrophoresis unit
-
Gel dock system
-
Tris-acetate-EDTA (TAE) buffer (1X)
-
Ultraviolet (UV) transilluminator
-
Ethidium bromide
-
Measuring cylinders
-
Microwave oven
-
Tris base
-
EDTA
-
Glacial acetic acid
-
Base pair ladder (100 bp)
-
Electrophoresis buffer (prepared from EDTA, Tris Base, Glacial acetic acid, mentioned below)
-
Dye (6X).
Preparation of 0.5 M ethylenediaminetetraacetic acid
For 1 L of solution, add 186.1 g of EDTA in 1000 ml of PCR grade, mix it well with the help of magnetic stirrer. Set the pH at 8.0. For pH adjustments add NaOH pellets.
PREPARATION OF STOCK SOLUTION AND DILUTION
For 1000 ml 50X TAE [Table 1]

-
Weigh 242 g Tris Base, transfer it to 2 ml beaker and add 750 ml Milli-Q water. Mix until it get dissolved
-
Add 100 ml of 0.5 M EDTA solution and 57.1 g glacial acetic acid. Mix well and set the pH at 8.3. Adjust the volume to 1000 ml by adding Milli-Q water
-
Filter to remove the un-dissolved material. Transfer it to autoclave bottle
-
Autoclave it at 15 lbs at 121°C for 20 min.
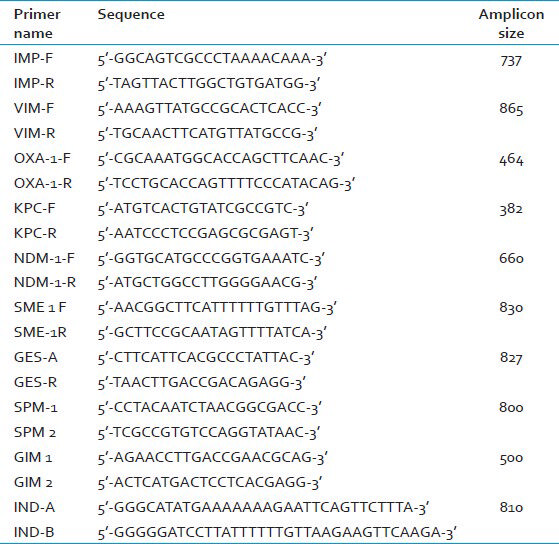
DNA is extracted from bacterial colonies using DNA extraction kits (QIAGEN). The presence of blaIMP, blaVIM, blaOXA, blaKPC, blaGES, blaSPM, blaGIM, blaIND, blaSME and blaNDM is detected by PCR. The primers are detailed in Table 2.[7891011121314]
For each target gene, PCR amplification is carried out in a 50 μl reaction volume. The reaction mixture contains 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 2 mM MgCl2, a 0.2 mM concentration of each dnTps, a 0.2 μM concentration of each specific primer, and 1.35 U of Taq DNA polymerase. For PCRs, an initial 10 min denaturation step at 95°C is performed followed by 35 cycles of 45 s of denaturation at 94°C, 45 s of primer annealing at respective temperature, and 50 s of primer extension at 72°C. Following the last cycle, an additional 7 min extension step is performed at 72°C, and the products are held at 4°C.
The cycling conditions have to be standardized in the laboratory for each gene. After agarose gel electrophoresis with ethidium bromide, the PCR products are to be analyzed under UV light.
Quality control
Escherichia coli ATCC 25922 (negative control) and K. pneumoniae ATCC 700603 (positive control) should be run as quality control. For each batch of multidrug-resistance, one batch of quality control is run. If the controls do not show the expected pattern the results of the samples plated on that day is considered as invalid.
Detection of NDM 1
NDM – 1 polymerase chain reaction
The presence of blaNDM-1 is detected by performing PCR, as described above using the primers listed in Table 2.[814]
Sequencing of blaKPC gene
For sequencing, the PCR is done as described above. Amplified products of the expected size are confirmed as KPC by sequencing. For typing of the blaKPC gene, overlapping PCR reactions will be performed using the following primer pairs:[15]
-
F-5’CGGAACCATTCGCTAAACTC3’ R-3’GGCGGCGTTATCACTGTATT5’
-
F-5’CGCCGTGCAATACAGTGATA3’ R-3’CGTTGACGCCCAATCC5’.
Amplification products are purified and sequencing is performed with an automated sequencer (ABI Prism 310; Applied Biosystems). Sequences are aligned and compared using the National Center for Biotechnology Information database.
Sequencing of blaNDM gene
Polymerase chain reaction of NDM-1 gene is performed as described above. Sequencing of the NDM-1 gene is done by using the primers:[15]
-
NDM-A: 5’- CACCTCATGTTTGAATTCGCC;
-
NDM-B: 5’- CTCTGTCACATCGAAATCGC (product size 984 bp).
The amplification products are sequenced with an automated sequencer. Sequences should be aligned and compared using the National Center for Biotechnology Information database.
Sequencing of OXA genes
For this, genomic DNA is extracted. PCR mixtures containing 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4) 2SO4, 2 mM MgSO4, nuclease-free bovine serum albumin (0.1 mg/ml), 0.1% Triton X-100, 1.5 mM MgCl2, 800 μM PCR nucleotide mix, and 1.25 U of Pfu DNA polymerase is prepared in a total volume of 50 μl.
Primers are used to amplify the entire sequence under the following conditions: 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 48°C for 40 s, and 72°C for 3 min and then 72°C for 6 min. Following purification using a PCR purification kit, the products are sequenced with an automated sequencer (ABI Prism 310; Applied Biosystems). The resulting sequences are analyzed using the online BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) software.[16]
ACKNOWLEDGMENTS
This study was funded by a grant from the Indian Council of Medical Research, Project code: I-800. We acknowledge the financial support of ICMR for the performance of this study.
Source of Support: This study was funded by a grant from the Indian Council of Medical Research, Project code: I-800. We acknowledge the financial support of ICMR for the performance of this study.
Conflict of Interest: None declared.
REFERENCES
- Extended-spectrum-beta-lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol. 2010;48:1019-25.
- [Google Scholar]
- Prevalence of multidrug resistance, ESBL and MBL production in Acinetobacter spp. Int J Recent Trends Sci Technol. 2013;6:139-43.
- [Google Scholar]
- Phenotypic and genotypic detection of metallo-beta-lactamases in imipenem-resistant Acinetobacter baumannii isolated from a Tertiary Hospital in Alexandria, Egypt. Res J Microbiol. 2011;6:750-60.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing, 20th Informational Supplement, January and June, 2010. Wayne, PA, USA: CLSI; 2010. M100-S 21
- [Google Scholar]
- Detection of metallo betalactamase producing Pseudomonas aeruginosa in hospitalized patients. Indian J Med Res. 2005;122:148-52.
- [Google Scholar]
- Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. 2011;104:40-5.
- [Google Scholar]
- New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg Infect Dis. 2011;17:103-6.
- [Google Scholar]
- SME 3, a novel member of the Serratia marcescens SME family of carbapenem-hydrolyzing beta-lactamases. Antimicrob Agents Chemother. 2006;50:3485-7.
- [Google Scholar]
- Emergence of KPC-producing Klebsiella pneumoniae in Italy. BMC Res Notes. 2010;3:40.
- [Google Scholar]
- Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla (OXA-51-like) genes. J Clin Microbiol. 2010;48:2476-83.
- [Google Scholar]
- Nosocomial spread of ceftazidime-resistant Klebsiella pneumoniae strains producing a novel class a beta-lactamase, GES-3, in a neonatal intensive care unit in Japan. Antimicrob Agents Chemother. 2004;48:1960-7.
- [Google Scholar]
- Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob Agents Chemother. 2004;48:4654-61.
- [Google Scholar]
- Identification and characterization of a new metallo-beta-lactamase, IND-5, from a clinical isolate of Chryseobacterium indologenes. Antimicrob Agents Chemother. 2007;51:2988-90.
- [Google Scholar]
- Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005--the new ‘MRSAs’? J Antimicrob Chemother. 2008;62:978-85.
- [Google Scholar]
- Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597-602.
- [Google Scholar]





