Translate this page into:
Diagnostic Performance of Multiplex PCR for Detection of Mycobacterium tuberculosis Complex in Presumptive Pulmonary Tuberculosis Patients and Its Utility in Smear Negative Specimens
Address for correspondence: Sarman Singh, MBBS, MD, All India Institute of Medical Sciences Bhopal, Medical College Building, AIIMS Bhopal Campus, Saket Nagar, Bhopal, 462020, Madhya Pradesh, India (e-mail: sarman_singh@yahoo.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
The primary objective of this study was to assess the diagnostic performance of multiplex polymerase chain reaction (mPCR) for the detection of Mycobacterium tuberculosis complex (MTBC) in presumptive pulmonary TB patients, in the setting of a tertiary level teaching hospital in central India, in comparison to liquid culture using BACTEC mycobacteria growth indicator tubes (MGIT) 960 TB system as the gold standard. The secondary objective was to assess the performance of mPCR for Ziehl Neelsen smear negative samples and ascertain the utility of this assay in smear negative samples.
Materials and Methods
Sputum or bronchoalveolar lavage samples were collected from patients who were adults, aged 18 years or older, presenting with presumptive pulmonary TB, and subjected to three microbiological investigations, that is, Ziehl Neelsen staining, mycobacterial culture using mycobacterial growth indicator tubes in the BD BACTEC MGIT 960 instrument, and the mPCR.
Statistical Analysis
For statistical analysis, 2 × 2 contingency tables were prepared and analyzed separately for all samples and for smear-negative samples using GraphPad and MedCalc tools. Sensitivity, specificity, positive predictive value, and negative predictive value (NPV) of mPCR were calculated by taking MGIT culture as the reference standard.
Results
For all samples (n = 114), sensitivity of mPCR for the detection of (MTBC) was 93.48% (95% confidence interval [CI]: 82.10–98.63%), specificity was 95.59% (95% CI: 87.64–99.08%), positive predictive value (PPV) was 93.48% (95% CI: 82.54–97.75%), and NPV was 95.59% (95% CI: 87.87–98.48%). For smear negative samples (n = 80), sensitivity was 80.00% (95% CI: 51.91–95.67%), specificity was 98.46% (95% CI: 91.72–99.96%), PPV was 92.31% (95% CI: 62.80–98.84%), and NPV was 95.52% (95% CI: 88.57–98.33%).
Conclusion
In this study, we were able to demonstrate the good performance characteristics of the mPCR for the detection of MTBC from clinical samples of patients with presumptive pulmonary tuberculosis, with MGIT liquid culture as the reference standard. It may be concluded that mPCR can be considered equivalent to MGIT culture in terms of clinical decision making and yield of positivity, owing to the good sensitivity and specificity for the detection of MTBC.
Keywords
Mycobacterium tuberculosis
multiplex PCR
molecular diagnosis of tuberculosis
Introduction
Tuberculosis (TB), caused by members of Mycobacterium tuberculosis complex, remains one of the most important causes of morbidity and mortality worldwide, with approximately 10 million new cases and 1.4 million deaths every year.[1] India bears 26% of this burden, that is, approximately 2.69 million cases every year.[2] The most common presentation is pulmonary TB, accounting for approximately 85% of all tubercular infections.[1]
The effective control and eventual elimination of TB depends on rapid diagnosis and treatment initiation. Rapid and accurate diagnosis is essential not only for better individual patient outcome but also from a public health perspective. To facilitate this, the World Health Organization (WHO) has recommended the use of molecular assays as initial tests for suspected TB patients and recommended commercial assays for this purpose.[3] These commercial assays require an initial setup of the specific analysis platform and continuous supply of assay kits. In such conditions, sputum smear microscopy is often used as the initial test. Although it is highly specific, it may be variably sensitive (sensitivity ranging from 20–80%).[4] Therefore, smear-negative samples need to be examined with another method; otherwise a substantial number of TB cases may remain undiagnosed and untreated. Culture isolation of bacilli remains the gold standard for diagnosis of TB because of its highest sensitivity, but its utility in clinical setting is limited by long turnaround time to positive result.[4]
The modality evaluated in this study is a multiplex polymerase chain reaction (PCR) assay developed and validated by K. Gopinath and S. Singh.[5] It uses specific gene targets to detect and differentiate M. tuberculosis complex and non-tuberculosis mycobacteria (NTM) directly from clinical samples. The sensitivity of the multiplex PCR in mycobacteriologically confirmed human immunodeficiency virus (HIV)-positive cases and HIV-negative cases was 97.1 and 97.05%, respectively, while the specificity was found to be 94.87% in both.[5]
This test was chosen as it is capable of simultaneous detection and differentiation of genus Mycobacteria, M. tuberculosis complex, and M. avium complex directly from a single clinical sample. Moreover, it does not require a specific platform for the assay, rather it can be performed in any laboratory equipped for molecular diagnostics. The turnaround time is significantly lower than solid or liquid culture and excellent performance for the detection of M. tuberculosis complex has been reported by the original authors.[5] This may be of particular significance in immunocompromised patients and in patients with specific risk factors, in whom nontubercular mycobacterial lung disease may be mistaken for pulmonary TB due to their similar clinical presentation, or it may be misdiagnosed (various commercial Nucleic Acid Amplification tests [NAAT]), or missed altogether (GeneXpert MTB/RIF).
In this study, we have assessed the diagnostic performance of multiplex PCR for the detection of M. tuberculosis complex in presumptive pulmonary TB patients, in the setting of a tertiary level teaching hospital in central India, in comparison to liquid culture using BACTEC mycobacteria growth indicator tubes (MGIT) 960 TB system as the gold standard. We have further assessed the performance of multiplex PCR for Ziehl Neelsen smear-negative samples, to ascertain the usefulness of this assay in smear-negative samples. Our secondary objective was to explore the prevalence of non-TB mycobacteria in patients presenting at this institute of central India.
Materials and Methods
A hospital-based cross sectional study was conducted in the Department of Microbiology, Pulmonary Medicine and General Medicine at a tertiary care hospital in central India from November 2019 to July 2021, after due ethical clearance was obtained from Institutional Ethics Committee. Optimal sample size for the study was calculated to be approximately 100 participants, using OpenEpi software. The formula used was Sample size n = [DEFF × Np(1 − p)]/ [(d2/Z21 − α/2 × (N − 1) + p × (1 − p)].
During the study period, on 2 days of each week, patients presenting to the outpatient department (OPD) with presumptive pulmonary TB were screened according to the inclusion and exclusion criteria by reviewing the OPD records. From among the patients who fulfilled all these criteria, recruitment was done in consecutive manner prospectively.
All patients provided written informed consent for participation and patients' confidentiality was maintained throughout the study. Samples were collected from patients who were adults aged 18 years or older presenting with presumptive pulmonary TB as per the definition laid down in the National Tuberculosis Elimination Program, or other patients aged 18 years or older presenting for screening of TB (e.g., contacts of microbiologically confirmed TB cases, people with HIV, diabetics, malnourished, cancer patients, patients on immunosuppressant or steroid therapy). Patients who declined to provide written informed consent or were currently on anti-TB therapy (ATT) or having history of administration of ATT within the past 6 months (from date of enrolment) were excluded from this study. Sputum samples were collected in accordance with the procedure recommended by the National Tuberculosis Elimination Program.[6] Bronchoalveolar lavage fluid samples were collected as per the procedure recommended by the American Thoracic Society Bronchoalveolar Lavage Guidelines[7] from patients who were unable to provide expectorated or induced sputum samples. The recruitment process and study procedures are summarized in ►Fig. 1.
- Flowchart of patient recruitment and study procedures. ATT, antituberculosis therapy; PCR, polymerase chain reaction.
Sputum samples were processed using a combination of N-acetyl-L-cysteine, a mucolytic agent, and sodium hydroxide, a decontaminating agent, under proper biosafety precautions, following the procedure reported previously.[8]
Smears were prepared from processed samples and stained using the Ziehl Neelsen staining method using 1% filtered carbol fuchsin as primary stain, 25% sulfuric acid as decolorizer, and 0.1% methylene blue as counterstain.[6] The prepared smears were then observed under 1000× magnification with oil immersion lens.[9]
For mycobacterial culture, all samples were inoculated into MGIT for culture isolation in the BD BACTEC MGIT 960 instrument (Becton Dickinson and Company, Sparks, Maryland, United States), following the procedures laid out by the manufacturer.[10]
For multiplex PCR assay, first, mycobacterial DNA was isolated from processed samples using the chloroform isoamyl alcohol method.[11]
The PCR master mix was prepared and PCR was performed as per previously published protocols.[5] The reaction mixture (25.5 μL) consisted of distilled water (13.7 μL), buffer (2.5 μL), dNTPs (4 μL), hsp forward primer (0.5 μL), hsp reverse primer (0.5 μL), ESAT-6 forward primer (0.5 μL), ESAT-6 reverse primer (0.5 μL), ITS MAC forward primer (0.5 μL), ITS MAC reverse primer (0.5 μL), Taq. Pol (0.3 μL), and template DNA (5 μL). The primer sequences and product lengths are summarized in ►Table 1. One negative template control and two positive controls (previously isolated DNA of M. tuberculosis H37Rv and M. avium complex strain) were included in each run of PCR. DNA amplification was performed for 30 cycles after a hot start for 10 minutes at 94°C. Each cycle consisted of denaturation for 1 minute at 94°C, annealing for 1 minute at 60°C and extension for 1 minute at 72°C. At the end, 10 minutes were allowed for final extension at 72°C.
| S. No. | Name | Sequence | Product size |
|---|---|---|---|
| 1 | ESAT-6 F | GCG GAT CCC ATG ACA GAG CAG CAG TGG A | 320bp |
| 2 | ESAT-6 R | CCC AAG CTT CCT ATG CGA ACA TCC CAG TGA CG | 320bp |
| 3 | ITS MAC F | CCC TGA GAC AAC ACT CGG TC | 144bp |
| 4 | ITS MAC R | ATT ACA CT TTC GAT GAA CGC | 144bp |
| 5 | hsp-65 F | ACC AAC GAT GGT GTG TCC AT | 441bp |
| 6 | hsp-65 R | CTT GTC GAA CCG CT ACC CT | 441bp |
Abbreviation: PCR, polymerase chain reaction.
After completion of PCR cycle, the amplified products were analyzed by electrophoresis in ethidium bromide (5 µg/mL)-stained 1.8% (w/v) agarose gels and photographed using gel documentation system. ►Fig. 2 shows the results of one batch of samples processed in the above manner.

- Visualization of results of multiplex polymerase chain reaction one batch of samples by agarose gel electrophoresis. (L, Ladder; NTC, negative template control; PC1, positive control with Mycobacterium tuberculosis H37Rv; PC2, positive control with Mycobacterium avium complex; S1 to S27—clinical specimens; Mycobacterium tuberculosis complex detected in: S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S20, S22, S23, S24, S25, S26, S27; No mycobacteria detected in: S19, S21).
For statistical analysis, 2 × 2 contingency tables were prepared and analyzed separately for all samples and for smear-negative samples using GraphPad and MedCalc tools. Sensitivity, specificity, positive predictive value, and negative predictive value of multiplex PCR were calculated by taking MGIT culture as the reference standard, and p-value was calculated with Fisher's exact test. Quantification of agreement was done with Kappa results. Diagnostic test characteristics were determined with 95% confidence intervals (CIs).
Results
In this study, total 114 samples were collected over a period of 14 months, out of which 102 (89.47%) were sputum samples and 12 (10.53%) were bronchoalveolar lavage fluid samples. The demographic and baseline clinical characteristics of the patients are summarized in ►Table 2.
| Characteristic | Number of samples (%) | |
|---|---|---|
| Sex | Female | 32 (28.07%) |
| Male | 82 (71.93%) | |
| Age | 18–45 years | 59 (51.75%) |
| 46–60 years | 33 (28.95%) | |
| 60 years + | 22 (19.30%) | |
| State of residence | Bihar | 1 (0.88%) |
| Madhya Pradesh | 109 (95.61%) | |
| Uttar Pradesh | 4 (3.51%) | |
| Presenting symptoms | Cough with expectoration | 78 (68.42%) |
| Fever | 46 (40.35%) | |
| Loss of appetite | 39 (34.21%) | |
| Weight loss | 33 (28.95%) | |
| Dry cough | 28 (24.56%) | |
| Dyspnea | 23 (20.18%) | |
| Other symptoms | 21 (18.42%) | |
| Chest pain | 14 (12.28%) | |
| Hemoptysis | 11 (9.65%) | |
| Altered consciousness | 2 (1.75%) | |
| HIV status | Negative | 61 (53.51%) |
| Positive | 2 (1.75%) | |
| Not tested | 51 (44.74%) | |
| Sample obtained | Bronchoalveolar lavage fluid | 12 (10.53%) |
| Sputum | 102 (89.47%) | |
Abbreviation: HIV, human immunodeficiency virus.
Taking all three modalities into account, M. tuberculosis complex was detected in 50 out of 114 specimens, giving a positivity rate of 43.86% for the entire specimen pool of the study. To elaborate further, out of 12 bronchoalveolar lavage (BAL) fluid samples, 3 were positive for M. tuberculosis complex and 9 were negative. However, out of 112 sputum samples, 47 were positive and 55 were negative. The detailed description of results of detection of M. tuberculosis complex from the two types of samples is depicted in ►Fig. 3. Thus, the positivity rate for BAL fluid samples was 25%, while for sputum samples the positivity rate was 46%.
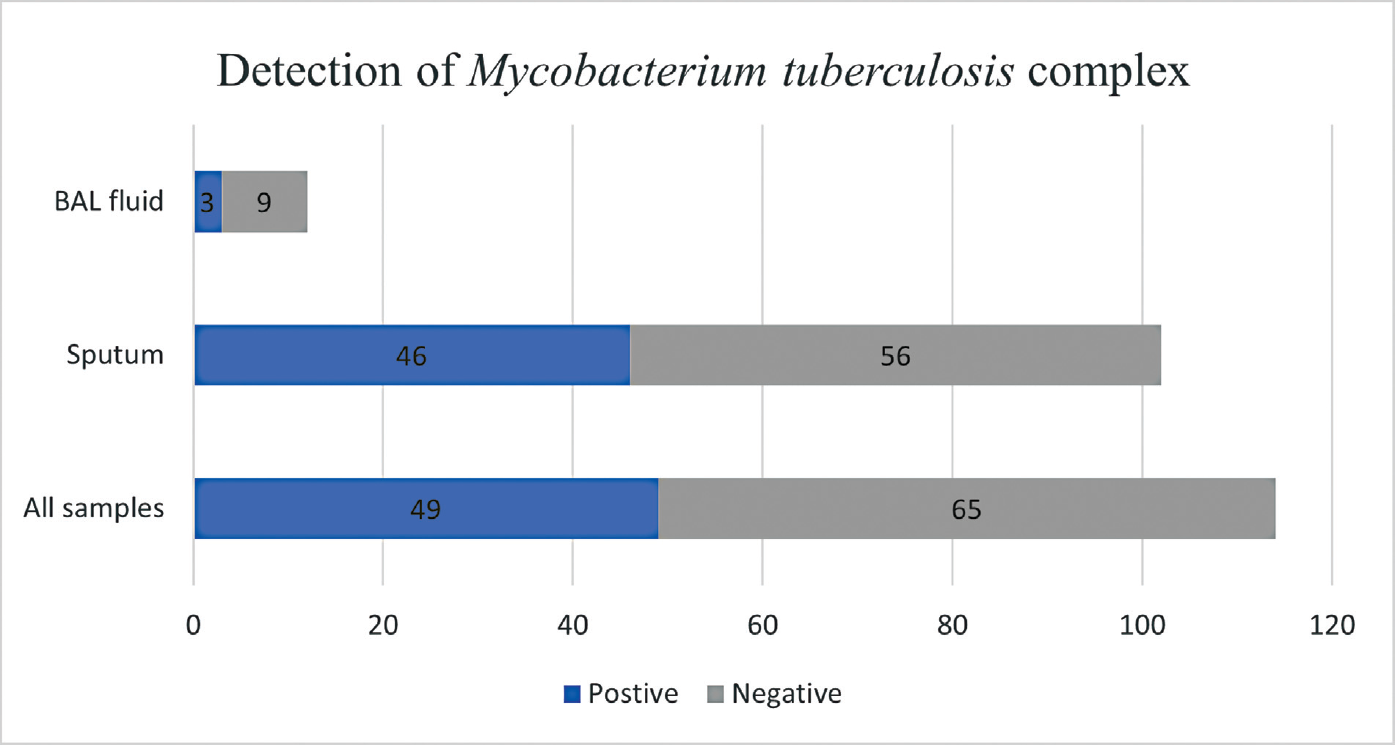
- Number of samples (total, BAL fluid, and sputum) testing positive and negative for Mycobacterium tuberculosis complex. BAL, bronchoalveolar lavage.
Positivity rate for the detection of M. tuberculosis complex was slightly higher in males (43.90%) as compared with females (43.75%). Among the three age groups, the positivity rate for the detection of M. tuberculosis complex was highest in the 18 to 45 years age group (50.85%), followed by the 46 to 60 years age group (39.39%) and was lowest in the 60+ years age group (31.82%). Among the 49 positive samples, the highest number belonged to male patients from the 18 to 45 years age group (19 samples or 38.78%), followed by females from the same age group and males from the 45 to 60 years age group (10 samples or 20.41% each).
Results of Ziehl Neelsen Staining: Out of the total sputum specimens (n = 102), 32 were positive for acid fast bacilli and 70 were negative. Among the positive specimens, 6 were graded 3 +, 15 were graded 2 +, 8 were graded 1 +, and 3 were graded scanty. Out of the total BAL specimens (n = 12), 10 were positive for acid fast bacilli and 2 were negative.
Results of MGIT Liquid Culture: Out of the total sputum specimens (n = 102), 43 were culture positive for M. tuberculosis complex and 59 were negative. Out of the total BAL specimens (n = 12), 3 were culture positive for M. tuberculosis complex and 9 were negative. Overall, 46 specimens were culture positive for M. tuberculosis complex and 68 were negative.
Results of Multiplex PCR: Out of 102 sputum specimens, 43 were positive for M. tuberculosis complex with the multiplex PCR and 59 were negative, while out of 12 BAL specimens, 9 were positive for M. tuberculosis complex with the multiplex PCR and 3 were negative. Overall, 46 specimens were multiplex PCR positive for M. tuberculosis complex and 68 were negative. Importantly, no non-TB mycobacterial species were detected in any of the specimens with the multiplex PCR.
A comparative description of the results of all three tests for M. tuberculosis complex for all samples is shown in ►Table 3. Overall, there were 19 samples with discordant results among the three tests, while there were 6 samples showing discordance between multiplex PCR results and MGIT liquid culture results.
| ZN Smear Result | mPCR Result | MGIT liquid culture | No. of samples (%) |
|---|---|---|---|
| Neg | Neg | Neg | 64 (56.14%) |
| Neg | Neg | Pos | 3 (2.63%) |
| Neg | Pos (MTB) | Neg | 1 (0.88%) |
| Neg | Pos (MTB) | Pos | 12 (10.53%) |
| Pos | Neg | Neg | 1 (0.88%) |
| Pos | Pos (MTB) | Neg | 2 (1.75%) |
| Pos | Pos (MTB) | Pos | 31 (27.19%) |
| Total | 114 (100.00%) | ||
Abbreviations: MGIT, mycobacteria growth indicator tube; mPCR, multiplex polymerase chain reaction; MTB, Mycobacterium tuberculosis complex; Neg, negative; Pos, positive; ZN, Ziehl Neelsen.
To fulfil the primary objective, the results of multiplex PCR for the detection of M. tuberculosis complex from all samples (n = 114) were compared with MGIT liquid culture as the reference standard using a 2 × 2 table. The sensitivity was found to be 93.48% (95% CI: 82.10–98.63%), the specificity was 95.59% (95% CI: 87.64–99.08%), positive predictive value was 93.48% (95% CI: 82.54–97.75%), and negative predictive value was 95.59% (95% CI: 87.87–98.48%). Accuracy was 94.74% (95% CI: 88.90–98.04%). Kappa was calculated to be 0.891 (standard error 0.043; 95% CI: 0.806–0.976), signifying almost perfect agreement. The two-tailed p-value calculated by Fisher's exact test was less than 0.0001; thus, these findings are statistically significant.
To fulfil the secondary objective, the results of multiplex PCR for the detection of M. tuberculosis complex from smear-negative samples (n = 80) were compared with MGIT liquid culture as the reference standards using a 2 × 2 table. The sensitivity was found to be 80.00% (95% CI: 51.91–95.67%), the specificity was 98.46% (95% CI: 91.72–99.96%), positive predictive value was 92.31% (95% CI: 62.80–98.84%), negative predictive value was 95.52% (95% CI: 88.57–98.33%). Accuracy was 95.00% (95% CI: 87.69–98.62%). Kappa was calculated to be 0.827 (standard error 0.084; 95% CI: 0.663–0.991), signifying almost perfect agreement. The two-tailed p-value calculated by Fisher's exact test was less than 0.0001; thus, these findings are statistically significant.
The diagnostic performance of multiplex PCR for all samples and for smear-negative samples is summarized in ►Table 4.
| Statistic | Value (95% CI) for all samples | Value (95% CI) for smear negative samples |
|---|---|---|
| Sensitivity | 93.48% (82.10–98.63%) | 80.00% (51.91–95.67%) |
| Specificity | 95.59% (87.64–99.08%) | 98.46% (91.72–99.96%) |
| Positive predictive value | 93.48% (82.54–97.75%) | 92.31% (62.80–98.84%) |
| Negative predictive value | 95.59% (87.87–98.48%) | 95.52% (88.57–98.33%) |
| Accuracy | 94.74% (88.90–98.04%) | 95.00% (87.69–98.62%) |
| Kappa | 0.891 (0.806–0.976) | 0.827 (0.663–0.991) |
| p-Value | < 0.0001 | < 0.0001 |
Abbreviations: CI, confidence interval; PCR, polymerase chain reaction
Discussion
In the current era, molecular methods of diagnosis have an undeniable and unequivocal role in TB diagnosis, due to their rapidity and accuracy. Although the commercially available platforms recommended by the WHO possess good performance characteristics, their large-scale implementation has been restricted in low-resource settings.[12] In such settings, development of molecular assays provides a more economical and practically feasible alternative since the assay can be customized as per the local infrastructure and human resources.
In the current study, a total of 114 samples were collected out of which 102 were sputum samples and 12 were BAL fluid samples. Low number of BAL fluid samples can be attributed to the change in hospital policy for bronchoscopy due to the ongoing COVID-19 pandemic. At the onset of the pandemic, bronchoscopy as a routine procedure was suspended, and it was only performed for limited indications.
The positivity rates for the detection of M. tuberculosis complex in this study were largely in accordance with the nationally and internationally reported data, in terms of age and sex distribution. Worldwide, adult men, women, and children account for 56, 32, and 12% cases, respectively.[1] Similarly, in India, adult males, females, and children account for 61.7, 32.65, and 5.65% cases of TB cases, respectively.[2] Majority (109 out of 114, or 95.61%) of the patients are residents of the central Indian state of Madhya Pradesh. Remaining (< 5%) are from neighboring states. This distribution indicates that the patients presenting to our hospital are from central India, and the findings of this study would likely be a true representation of the local population.
In this study, we have been able to demonstrate good sensitivity and specificity of multiplex PCR for the detection of M. tuberculosis complex from pulmonary samples, regardless of their smear results. The sensitivity (93.48% [95% CI: 82.10–98.63%]) and specificity (95.59% [95% CI: 87.64–99.08%]) of multiplex PCR (with MGIT culture as the reference) from this study were fairly good, considering the significant reduction in terms of time taken to positive result by multiplex PCR as compared with MGIT culture. Even for smear-negative samples, we have been able to demonstrate satisfactory diagnostic performance of multiplex PCR, with sensitivity of 80.00% (95% CI: 51.91–95.67%), specificity of 98.46% (95% CI: 91.72–99.96%), positive predictive value of 92.31% (95% CI: 62.80–98.84%), and negative predictive value of 95.52% (95% CI: 88.57–98.33%). Thus, the assay is a potentially valuable tool to diagnose smear-negative and paucibacillary samples rapidly and accurately.
These findings are supported by other recently published reports. For example, one report from Meriki et al[13] found the sensitivity and specificity of duplex PCR assay for all samples to be 93.5 and 94%, respectively; and for smear-negative culture positive samples, the sensitivity was 87.5%. Furthermore, a systematic review and meta-analysis to assess the overall accuracy of RT-PCR assay for TB diagnosis in different samples for individuals with active pulmonary and extrapulmonary infection[14] reported pooled sensitivity of 0.96 and pooled specificity of 0.92.
There were 19 samples which have discordant results. Most of them (n = 12) were samples that were smear-negative, but tubercle bacilli were detected by both multiplex PCR and MGIT liquid culture. This is not surprising, given the variable sensitivity of smear microscopy for the detection of acid-fast bacilli. Next, there were three samples that were smear-negative, as well as multiplex PCR negative, liquid culture grew M. tuberculosis complex. It is possible that the bacillary load was too low to be detected by multiplex PCR, or there were some unrecognized PCR inhibitors in the samples.[13] These samples reiterate the role of culture as an important part of TB diagnosis.
In two samples that were smear-positive, and M. tuberculosis complex was detected by multiplex PCR, but the MGIT liquid culture failed to show any growth; and there was one sample that was smear-positive but M. tuberculosis complex was not detected by either multiplex PCR or liquid culture. The presence of dead bacilli due to previously treated TB in these samples was ruled out by reviewing clinical records of these patients. As per previous reports, this discordance may be attributed to presence of nonculturable M. tuberculosis, which has low viability making them impossible to culture.[15,16]
Furthermore, there was one sample that was smear-negative, multiplex PCR positive, and liquid culture negative. Again, the presence of dead bacilli due to past treatment of TB was ruled out by review of clinical records. One of the possible reasons behind negative culture result may be excessive harshness of the decontamination procedure used for the concerned specimen. It has also been reported in existing literature that culture recovery of mycobacteria from salivary sputum specimens reduces after centrifugation due to the low buoyant density of bacilli, which might also be another possible reason for negative culture.[17]
In this study, no other mycobacterial species apart from M. tuberculosis complex was detected by multiplex PCR or by liquid culture. This was a major limitation as we were not able to assess the diagnostic performance of the multiplex PCR for detection and differentiation of non-TB mycobacterial species, or its practical utility in laboratory diagnosis of non-TB mycobacteria in suspected TB patients and subsequent impact on treatment. Detection of NTM from a range of clinical samples using multiplex PCR has been previously reported from several studies in North India. One study[18] from North India reported detection of NTM in 24 out of 572 clinical samples, while another study[19] from the same region reported detection of NTM in 13 out of 436 clinical samples. Other modalities have also been used for the detection of NTM, for example, one study[20] using The GenoType Mycobacterium common mycobacteria/additional species assay (Hain Lifescience, Nehren, Germany) found that out of 1,080 clinical samples, NTM was detected in 60 samples.
One of the plausible reasons for lack of detection of NTM in the present study could be the inadequate sample size due to low prevalence of NTM in this particular geographical region. In previous reports, a wide range of NTM prevalence has been reported from various parts of India,[21] but there has been only one study that reported NTM prevalence of 1.3% from sputum specimens of suspected TB patients in Madhya Pradesh. Most common species identified were M. abscessus and M. intracellulare.[22] Studies with larger and more diverse participant pool, along with wider geographical coverage, are needed to fully illustrate the prevalence and species distribution of NTM in central India. Inclusion of elderly females and extrapulmonary specimens may prove to be especially valuable for this purpose.
Overall, multiplex PCR can be considered equivalent to MGIT culture in terms of clinical decision making and yield of positivity with added advantage of rapidity. The most important advantages offered by multiplex PCR over MGIT culture are that of reduced turnaround time and simultaneous detection of other mycobacterial species. The maximum benefit of implementation of molecular assays can be achieved in the setting of tertiary level centers where the setup for molecular work already exists for other diagnostic or research work, or where such a setup is under development. Such centers usually cater to a large peripheral population, and there is availability of skilled human resources. Assays provide a more sustainable and worthwhile alternative to commercial assays because the reagents and equipment are not specific to one disease or organism, rather there is unlimited potential for customization of the assays and their application in diagnosis of other infectious diseases. Furthermore, dependency on commercial kits and consumables can also be reduced, and the interruption of commercial supplies would not affect patient care services.
Conclusion
In this study, we were able to demonstrate the good performance characteristics of the multiplex PCR for the detection of M. tuberculosis complex from clinical samples of patients with presumptive pulmonary TB, with MGIT liquid culture as the reference standard.
Multiplex PCR shows robust diagnostic performance with significant advantage over MGIT liquid culture in terms of turnaround time; hence, it can be considered equivalent to MGIT culture in terms of clinical decision making and yield of positivity, owing to the good sensitivity and specificity for the detection of M. tuberculosis complex.
Ethical Approval
This research study was approved by the Institutional Human Ethics Committee of All India Institute of Medical Sciences Bhopal, Letter of approval number IHEC-LOP/2019/ MD0100, dated 23 October 2019.
Authors' Contributions
Lonika Lodha performed the scientific work and prepared the manuscript draft. Shivkumar Rashmi Mudliar performed the scientific work and edited the manuscript draft. Jitendra Singh supervised the scientific work and edited the manuscript draft. Anand Maurya supervised the scientific work and edited the manuscript draft. Alkesh Kumar Khurana supervised the clinical data collection and approved the final manuscript. Sagar Khadanga supervised the clinical data collection and approved the final manuscript. Sarman Singh supervised the scientific work and approved the final manuscript.
Conflict of Interest
None.
Funding
This study was a part of the MD thesis work conducted by the author, which was supported by a financial grant from the Indian Council of Medical Research [Award Letter Number 3/2/Dec-2019/PG-Thesis-HRD (29); Dated: 23.03.2020]. The authors are grateful to ICMR for this incentive.
References
- Global Tuberculosis Report 2020. 2020 Accessed February 17, 2022 https://www.who.int/publications/i/item/9789240013131.
- [Google Scholar]
- Division. Indian TB Report 2020- Ministry of Health and Family Welfare. Published 2020. Accessed February 17, 2022 https://tbcindia.gov.in/showfile.php?lid=3538.
- [Google Scholar]
- WHO consolidated guidelines on tuberculosis. Module 3: diagnosis – rapid diagnostics for tuberculosis detection. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO 2020 Published online 2020. Accessed April 5, 2021. Accessed February 17, 2022 https://www.who.int/publications/i/item/9789240029415.
- [Google Scholar]
- Diagnosis of pulmonary tuberculosis: recent advances and diagnostic algorithms. Tuberc Respir Dis (Seoul). 2015;78(02):64-71.
- [CrossRef] [PubMed] [Google Scholar]
- Multiplex PCR assay for simultaneous detection and differentiation of Mycobacterium tuberculosis, Mycobacterium avium complexes and other Mycobacterial species directly from clinical specimens. J Appl Microbiol. 2009;107(02):425-435.
- [CrossRef] [PubMed] [Google Scholar]
- Technical and Operational Guidelines for TB Control in India 2016: Central TB Division. Cent Tuberc Div. Published online 2016. Accessed February 17, 2022 https://tbcindia.gov.in/index1.php?lang=1&level=2&sublinkid=4573&lid=3177
- [Google Scholar]
- Accessed February 17, 2022 https://www.thoracic.org/professionals/clinical-resources/video-lecture-series/bronchoscopy/.
- Nontuberculous mycobacterial infections in Indian AIDS patients detected by a novel set of ESAT-6 polymerase chain reaction primers. Jpn J Infect Dis. 2007;60(01):14-18.
- [Google Scholar]
- Laboratory Services in Tuberculosis Control: Culture. Published online 1998:97. Accessed February 17, 2022 http://apps.who.int/iris/bitstream/handle/10665/65942/WHO_TB_98.258_(part3).pdf?sequence=3.
- [Google Scholar]
- BD BACTEC MGIT 960 System for Mycobacteria Testing. Published online 2000:6. Accessed February 17, 2022 https://www.bd.com/BD_BACTEC-MGIT_PE_TR.
- [Google Scholar]
- TB Research Team. Diagnostic usefulness of Xpert MTB/RIF assay for detection of tuberculous meningitis using cerebrospinal fluid. J Infect. 2017;75(02):125-131.
- [CrossRef] [PubMed] [Google Scholar]
- The implementation of Xpert MTB/RIF assay for diagnosis of tuberculosis in Nepal: A mixed-methods analysis. PLoS One. 2018;13(08):e0201731. DOI: 10.1371/JOURNAL.PONE.0201731
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of the performance of an in-house duplex PCR assay targeting the IS6110 and rpoB genes for tuberculosis diagnosis in Cameroon. BMC Infect Dis. 2020;20(01):791.
- [CrossRef] [PubMed] [Google Scholar]
- Diagnostic accuracy of in-house real-time PCR assay for Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2019;19(01):701. DOI: 10.1186/S12879-019-4273-Z
- [CrossRef] [PubMed] [Google Scholar]
- Assessment of sputum smear-positive but culture-negative results among newly diagnosed pulmonary tuberculosis patients in Tanzania. Int J Gen Med. 2017;10:199-205.
- [CrossRef] [PubMed] [Google Scholar]
- The incidence and clinical implication of sputum with positive acid-fast bacilli smear but negative in mycobacterial culture in a tertiary referral hospital in South Korea. J Korean Med Sci. 2008;23(05):767-771.
- [CrossRef] [PubMed] [Google Scholar]
- GeneXpert MTB/RIF outperforms mycobacterial culture in detecting mycobacterium tuberculosis from salivary sputum. BioMed Res Int. 2018;2018 1514381 DOI: 10.1155/2018/1514381
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of an in-house multiplex PCR with two commercial immuno-chromatographic tests for rapid identification and differentiation of MTB from NTM isolates. Int J Mycobacteriol. 2014;3(01):50-56.
- [CrossRef] [PubMed] [Google Scholar]
- High prevalence of non-tuberculous mycobacterial disease among non-HIV infected individuals in a TB endemic country–experience from a tertiary center in Delhi, India. Pathog Glob Health. 2014;108(02):118-122.
- [CrossRef] [PubMed] [Google Scholar]
- Role of GenoType(®) Mycobacterium common mycobacteria/additional species assay for rapid differentiation between Mycobacterium tuberculosis complex and different species of non-tuberculous mycobacteria. J Lab Physicians. 2013;5(02):83-89.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology, diagnosis & treatment of non-tuberculous mycobacterial diseases. Indian J Med Res. 2020;152(03):185-226.
- [CrossRef] [PubMed] [Google Scholar]
- Public health relevance of non-tuberculous mycobacteria among AFB positive sputa. Germs. 2017;7(01):10-18.
- [CrossRef] [PubMed] [Google Scholar]






