Translate this page into:
Early Biochemical Markers in Predicting the Clinical Outcome of COVID-19 Patients Admitted in Tertiary Care Hospital
Address for correspondence: Suprava Patel, MD, Department of Biochemistry, All India Institute of Medical Sciences, Raipur, Chhattisgarh, 492099, India (e-mail: dr_suprava@yahoo.co.in).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction
An array of routinely accessible serum biomarkers was assessed to explore their overall impact on severity and mortality in coronavirus disease 2019.
Materials and Methods
A retrospective analysis of 1,233 adults was conducted. The study groups comprised 127 nonsurvivors and 1,106 survivors. Data for demographic details, clinical presentations, and laboratory reports were recorded from the medical record section. The predictors were analyzed for their influence on mortality.
Results
The mean (+ standard deviation) age of the patients in the nonsurvivor group was 58.8 (13.8) years. The mean age (56.4 years) was highest in severe grade patients. The odds ratio for death was 2.72 times for patients above the age of 40 years. About 46% of nonsurvivors died within 5 days of admission. Males were found to be more prone to death than females by a factor of 1.36. Serum urea depicted highest sensitivity (85%) for nonsurvival at 52.5 mg/dL. Serum albumin (3.23 g/dL), albumin-to-globulin ratio (0.97), and C-reactive protein-to albumin ratio (CAR) (2.08) showed a sensitivity of more than 70% for mortality outcomes. The high hazard ratio (HR) for deceased patients with hyperkalemia was 2.419 (95% confidence interval [CI] = 1.96–2.99; p < 0.001). The risk for nonsurvival was increased with elevated serum creatinine by 15.6% and uric acid by 21.7% (p < 0.001). The HR for hypoalbuminemia was 0.254 (95% CI: 0.196–0.33; p < 0.001) and CAR was 1.319 (95% CI: 1.246–1.397; p < 0.001). Saturation of oxygen (p < 0.001), lactate dehydrogenase (p = 0.006), ferritin (p = 0.004), hyperuricemia (p = 0.027), hyperkalemia (p < 0.001), hypoalbuminemia (p = 0.002), and high CAR values (0.031) served as potential predictors for mortality.
Conclusion
Adjusting for all the predictor variables, serum uric acid, potassium, albumin, and CAR values at the time of admission were affirmed as the potential biomarkers for mortality.
Keywords
total clinical severity score
serum markers
mortality predictor
sensitivity and specificity
primary biomarkers
Key Findings
Nearly 46% of nonsurvivors succumbed within 5 days of admission.
The probability for death was more than twice in patients above the age of 40 years.
Dyselectrolemia was significantly associated with the severity.
Uremia depicted the highest specificity for nonsurvival.
Hyperuricemia displayed considerable impact on mortality and significantly reduced the duration of hospitalization.
Hyperkalemia, hypoalbuminemia, and C-reactive protein-to albumin ratio (CAR) affirmed as independent predictor markers for nonsurvival.
Key Implications
Early assessment of disease progression by serum biomarkers is the critical factor for a better outcome.
Clinicians must understand the interplay of basic laboratory parameters in predicting mortality.
Serial monitoring of one or more predictive parameters is of paramount importance for early recovery.
Introduction
The World Health Organization (WHO) declared severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) as a pandemic outbreak owing to high mortality evidenced worldwide.[1] The severity was significantly associated with elderly individuals and those with underlying comorbid conditions like diabetes mellitus (DM), hypertension, chronic kidney disease, underlying cardiac diseases, sickle cell disease, asthma, chronic obstructive pulmonary disease, and many more.[2,3] Sudden onset of acute respiratory distress syndrome due to coronavirus disease 2019 (COVID-19) warrants early assessment of severity and intervention to reduce the mortality. Clinical judgment regarding the disease progression is complex and purely relies on blood test reports.
Therefore, blood investigations play a prime role in assessing the disease progression. Various blood parameters are defined for a poor outcome, such as serum ferritin, interleukin-6 (IL-6), procalcitonin, creatine kinase total, creatine kinase-MB (CK-MB), and blood D-dimer levels.[4-7] However, laboratory measurements of these parameters are not feasible in many small-scale laboratories (as in remote areas) and not affordable by many. In such a scenario, the laboratory outsources the samples to other laboratories and thus results in delayed reporting. By the time the clinician has the laboratory reports, the patients' condition might have deteriorated enough to revive. It would be a great help if the mortality outcome could be assessed by serially monitoring the basic blood parameters.
An array of routinely accessible blood biomarkers has been assessed in this study to observe their overall impact on mortality. These parameters can be tested in all laboratories, do not require highly sophisticated instruments, and are relatively inexpensive. Many government sector hospitals also provide the facility of these tests free of cost. An interplay of these parameters would provide considerable information regarding the disease prognosis at an early stage. Only a few articles have been published regarding biochemical changes in patients of COVID-19 worldwide. A serial increase in serum C-reactive protein (CRP) or lactate dehydrogenase (LDH) could warn regarding progression to cytokine storm.[8,9] Deranged liver enzymes or renal parameters could indicate inflammatory involvement in various organs and delayed recovery.[9-11] Dyselectrolemia could be an early indicator of disease severity.[12,13] Similarly, hypoproteinemia and hypoalbuminemia have been associated with adverse outcomes in admitted patients.[14,15]
A combination of markers provides better insight into the adverse outcome. Raised CAR is one of the vital predictor markers for systemic inflammation and high mortality in patients in intensive care units (ICUs).[16,17] However, the effectiveness in assessing mortality is not much reported.
The first and most easily accessible biochemical markers are the routine serum parameter testing that can assess the biochemical changes readily. These markers also aid in monitoring the progression of disease from mild-to-severe form and thus reduce the burden of healthcare facilities available by providing early diagnosis of this shift. Therefore, the treatment modality can be changed effectively by readily looking at these biochemical parameters and help in knowing the prognosis of the disease.
It is imperative to understand the influence of the primary biomarkers on mortality outcomes. Therefore, the study was aimed to explore the effect of serum biomarkers on survival in COVID-19 cases admitted to the institute.
Materials and Methods
Study Subjects and Data Collection
The study was conducted in a tertiary care hospital in the state. It was initiated after the institute ethics committee approved it, along with a consent waiver. It was a retrospective analysis of serum biomarkers in COVID-19 confirmed cases admitted to the institute. A total of 1,233 cases of more than 18 years old were enrolled for the study during the 4 months of study duration, and Helsinki's guidelines for good clinical practice were strictly applied. Data collection for demographic and clinical details was collected from the institute's medical record department. The investigators collected details of the patient's demography and clinical presentation from the medical record section of the institute. The laboratory parameters investigated within 24 hours of admission were noted. Only reverse transcription polymerase chain reaction confirmed cases were included in the study. Patients with incomplete data (clinical and laboratory reports), pregnant and lactating females, and who had blood transfusions in the last 12 weeks were excluded from the study.
The study participants were further grouped as survivors and nonsurvivors to estimate the mortality risk of the predictor variables. Patients' who recovered and were discharged from the hospital were grouped as survivors. Those who died of COVID-19 while undergoing treatment in the hospital were grouped as nonsurvivors. Based on percentage saturation of oxygen (SpO2%), the study population was grouped as mild (> 95%), moderate (90–94%), and severe (< 90%).[1]
After entering the completed clinical data, each of the subjects was assigned a comorbidity score as per the number of comorbid conditions associated. In addition to the comorbidity score, the numbers of clinical signs, symptoms, and survival status, the investigator assigned a total clinical severity (TCS) score to each of the patients.
The biochemical markers for renal function tests, liver function test, and the inflammatory markers (high-sensitivity-CRP [hs-CRP], LDH, and ferritin) were noted. Although not reported, the investigators calculated the following ratios for analysis purposes: serum glutamate-oxaloacetate transaminase-to-serum glutamate-pyruvate transaminase (SGOT/SGPT), albumin-to-globulin (AGR), CAR.
Serum hs-CRP, LDH, and ferritin were considered the inflammatory marker variables for analysis.
The demographic, clinical, and laboratory were considered as the predictors or the independent variables for the mortality outcome.
Statistical Analysis
The analysis was computed using IBM SPSS 20, and p-value less than 0.05 was used for statistical significance. The normality distribution of the continuous data was determined by the Shapiro–Wilk test, histogram, and Q-Q plots. The continuous variables were presented in both mean with standard deviation (SD) and median with interquartile range (IQR). Availability of both data subsets would be beneficial for readers better understand the distribution of the data in the study population. IQR denoted the first quartile (Q1 = 25th percentile) and the third quartile (Q3 = 75th percentile) values. Parametric independent samples t-test and the nonparametric Mann–Whitney U test as applicable for the respective data were used to compare the continuous variables between the survivor and nonsurvivor groups.
Receiver operating characteristics (ROC) with area-under-curve and cutoff values were calculated for the nonsurvival outcome. The Kaplan–Meier curve was performed to establish the significant relationship between the predictors with survival. Multiple cox regression analyses were performed to find out the survival function and hazard ratio (HR) of each independent predictor for mortality. The significant, influential predictors for nonsurvival after adjusting with other determinants were analyzed by multivariate cox-regression.
Results
Study Population and Frequency Distribution of Demographic Variables and Clinical Details in the Study Groups
The total number of survivors in the study population was 1,106 (89.7%), and that of the deceased was 127 (10.3%). The study population consisted of 69.2% (n = 853) males and 30.8% (n = 380) females.
As depicted in ►Fig. 1, almost 50% of the patients aged more than 60 years did not survive. Nearly 89% of nonsurvivors were above 40 years old, and nearly three-fourth (74.8%) were males. The odds ratio (OR) for nonsurvival was 2.72 times in patients above 40 years of age than their younger counterparts (95% confidence interval [CI]: 1.55–5.103; p < 0.001). The OR for death was 36% times more (OR; 95% CI: 1.36; 0.89–2.08; p = 0.15) in males than females. In nearly 75% of cases, the patients died within 10 days of admission, and almost 46% of nonsurvivors succumbed within 5 days of admission, while a median (IQR) of the duration of hospitalization was 6 (2–11) days. Of all the nonsurvivors, 71.7% got admitted under severe grade COVID-19 (SpO2 < 90%), and 35.4% had no associated comorbidities. The chances for nonsurvival were 11.88 times (95% CI: 7.84–18.01; p < 0.001) for severe cases compared with nonsevere ones. Similarly, the probability for death was increased by 47% in patients with some sort of comorbidity than those without so (95% CI: 1.003–2.156; p = 0.047). The OR for mortality was 3.61 times (95%CI: 1.74–7.5; p < 0.001) in the presence of multiple clinical symptoms. The majority of the nonsurvivors (63%) had a TCS score of less than or equal to 3, and the OR for nonsurvival was 1.58 (95% CI: 1.075–2.32; p = 0.019) in those with higher TCS score.
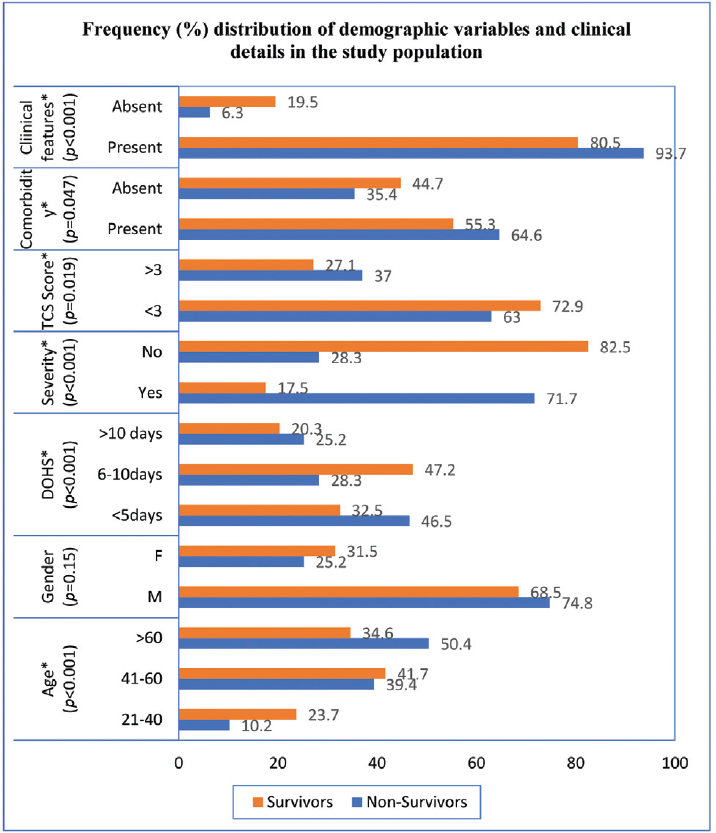
- Frequency distribution of demographic variables and clinical details in the study population (n = 1233). * denotes significance at p-value of 0.05; age depicted in years; M: males; F: females; DOHS: duration of hospitalization stay in days; TCS: total clinical severity score.
Of all the comorbidities, hypertension was the commonest with a frequency percentage of 45.6% in deceased patients, followed by DM in 41.7% (►Table 1). At the time of admission, breathlessness was the major presentation in 82.6% of nonsurvivors (►Fig. 2).
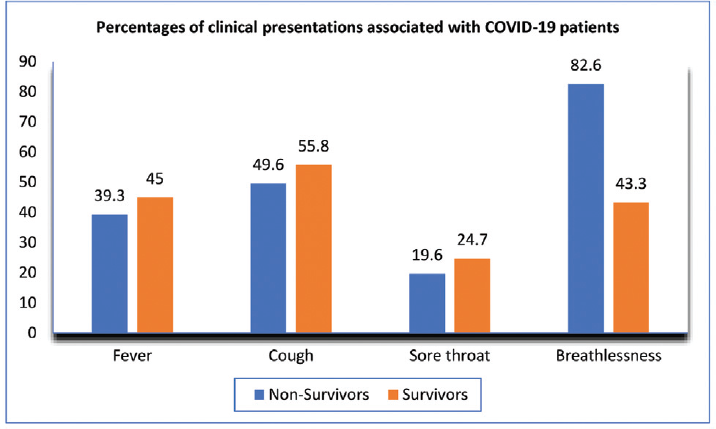
- Percentage distribution of clinical presentations associated with coronavirus disease 2019 (COVID-19) patients. The figures represent the percentage in each group.
| Comorbidities | Nonsurvivors N = 127 (10.3%) | Survivors N = 1,106 (89.7%) |
|---|---|---|
| n (%) | n (%) | |
| DM | 53 (41.7) | 390 (35.2) |
| Hypertension | 58 (45.6) | 409 (36.3) |
| CAD | 10 (7.8) | 67 (6.1) |
| CVA | 0 | 2 (0.1) |
| CKD | 7 (5.5) | 16 (1.4) |
| TB | 1 (0.7) | 13 (1.1) |
| Cancer | 0 | 7 (0.6) |
| SCD | 1 (0.7) | 3 (0.3) |
| Hyperthyroidism | 0 | 1 (0.1) |
| Hypothyroidism | 0 | 4 (0.4) |
| COPD | 2 (1.5) | 20 (1.8) |
| Others | 1 (0.7) | 5 (0.5) |
Abbreviations: CAD, coronary artery disease; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; COVID-19, coronavirus disease 2019; CVA, cerebrovascular accident; DM, diabetes mellitus; HTN, hypertension; n(%), denotes column percentage; SCD, sickle cell disease; TB, tuberculosis.
Comparison of Mean (SD) and Median (IQR) Values of the Study Variables between Survivors and Nonsurvivors
The mean (SD) age of the nonsurvivors was 58.8 (13.8) years that was significantly higher than the survivor cases (p = 0.02) (►Table 2). The median SpO2% in the study population was 95%, with a range of 63 (37–100). The median SpO2% at admission was significantly low in the nonsurvivor group (p < 0.001). The median duration of hospital stay was 6 days with a range of 24 (1–25) days for nonsurvivors, whereas it was 7 days with a range of 49 (0–49) days (p < 0.001). The TCS score did not report any substantial difference between the groups (p = 0.245). The median (IQR) SpO2 recorded in nonsurvivor cases at the time of admission was 80 (70–90), which was relatively low when compared with those recovered (p < 0.001).
| Variables (Units) | Study population, n = 1,233 | Nonsurvivors n = 127 (10.3%) | Survivors n = 1,106 (89.7%) | p-Value | |||
|---|---|---|---|---|---|---|---|
| Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | Mean (SD) | Median (IQR) | ||
| Age (y) | 53.5 (15.79) | 55 (42.5–65) | 58.8 (13.8) | 61 (50–69) | 52.84 (15.99) | 55 (42–65) | 0.02a |
| SpO2 (%) | 92 (8.96) | 95 (90–98) | 78.11 (13.69) | 80 (70–90) | 93.61 (6.59) | 96 (92–98) | < 0.001b |
| DOHS (d) | 8.06 (5.37) | 7 (5–10) | 6.98 (5.4) | 6 (2–11) | 8.19 (5.4) | 7 (5–10) | < 0.001b |
| Comorbidity score | 0.87 (0.91) | 1 (0–2) | 1.05 (0.94) | 1 (0–2) | 0.85 (0.9) | 1 (1–4) | 0.018b |
| TCS score | 2.6 (1.53) | 3 (1–4) | 2.92 (1.46) | 3 (2–4) | 2.57 (1.53) | 3 (1–4) | 0.245 |
| hs-CRP (mg/L) | 54.07 (59.65) | 29 (5–87) | 105.87 (65.8) | 96 (50–173) | 48.12 (55.93) | 21 (4–78) | < 0.001a |
| LDH (U/L) | 578.08 (298.43) | 511 (347–739) | 872.91 (345.77) | 835 (606–1128) | 544.23 (272.99) | 486.5 (331.5–688.3) | < 0.001a |
| Ferritin (ng/mL) | 453.88 (479.03) | 273 (99.5–644) | 985.19 (561.76) | 889 (520–1507) | 392.87 (428.59) | 237 (93.8–530.5) | < 0.001b |
| Urea (mg/dL) | 44.03 (43.92) | 30 (21–48) | 102.55 (73.62) | 76 (48–140) | 37.31 (33.07) | 28 (20–42) | < 0.001b |
| Creatinine (mg/dL) | 1.42 (1.64) | 1.1 (0.9–1.3) | 2.38 (2.68) | 1.3 (1.0–2.6) | 1.31 (1.4) | 1.1 (0.9–1.3) | < 0.001b |
| Uric acid (mg/dL) | 4.87 (2.21) | 4.5 (3.4–5.7) | 6.31 (3.62) | 5.3 (3.7–8.2) | 4.71 (1.93) | 4.4 (3.4–5.6) | < 0.001b |
| Na+ (mmol/L) | 138.19 (5.96) | 138 (135–141) | 142.17 (9.9) | 140 (135–145) | 137.73 (5.1) | 138 (135–141) | < 0.001a |
| K+ (mmol/L) | 4.19 (0.69) | 4.1 (3.8–4.5) | 4.68 (0.9) | 4.6 (4.0–5.4) | 4.13 (0.6) | 4.1 (3.8–4.5) | < 0.001a |
| Cl- (mmol/L) | 103.67 (5.5) | 104 (100–107) | 105.43 (9.4) | 103 (99–108) | 103.47 (4.8) | 104 (101–107) | < 0.001a |
| TBil (mg/dL) | 0.8 (0.83) | 0.63 (0.47–0.87) | 1.2 (1.6) | 0.82 (0.53–1.17) | 0.76 (0.7) | 0.62 (0.47-.83) | < 0.001b |
| DBil (mg/dL) | 0.25 (0.47) | 0.16 (24–53) | 0.51 (0.9) | 0.26 (0.16–0.43) | 0.22 (0.3) | 0.16 (0.11-.22) | < 0.001b |
| SGOT (U/L) | 54.24 (97.92) | 34 (24–53) | 114.03 (198.2) | 55 (34–92) | 47.37 (75.8) | 32 (24–49) | < 0.001b |
| SGPT (U/L) | 49.06 (83) | 31 (17–50) | 97.15 (187.7) | 42 (26–85) | 43.54 (58) | 29 (16–47) | < 0.001b |
| SGOT/SGPT | 1.34 (0.73) | 1.21 (0.88–1.67) | 1.49 (0.9) | 1.26 (0.84–1.91) | 1.33 (0.7) | 1.2 (0.9–1.6) | 0.28 |
| ALP (U/L) | 90.09 (61.88) | 75 (60–100) | 118.7 (84.4) | 105 (76–134) | 86.8 (57.9) | 74 (59–96) | < 0.001b |
| GGT (U/L) | 57.56 (73.38) | 36 (22–66) | 75.5 (61.1) | 54 (33–95) | 55.5 (74.4) | 35 (21–62) | < 0.001b |
| T. Protein (gm/dL) | 6.79 (0.85) | 6.84 (6.31–7.31) | 6.17 (0.9) | 6.2 (5.5–6.9) | 6.86 (0.8) | 6.9 (6.4–7.3) | 0.002a |
| Albumin (gm/dL) | 3.57 (0.62) | 3.62 (3.17–4.03) | 2.88 (0.5) | 2.9 (2.5–3.2) | 3.65 (0.6) | 3.7 (3.3–4.1) | 0.085 |
| Globulin (gm/dL) | 3.22 (0.62) | 3.15 (2.84–3.52) | 3.29 (0.7) | 3.3 (2.8–3.7) | 3.21 (0.6) | 3.1 (2.8–3.5) | 0.096 |
| AGR | 1.15 (0.29) | 1.12 (0.95–1.32) | 0.91 (0.24) | 0.88 (0.76–1.03) | 1.17 (0.28) | 1.16 (0.98–1.34) | < 0.001b |
| CAR | 1.71 (2.03) | 0.83 (0.14–2.69) | 3.83 (2.6) | 3.28 (1.6–5.8) | 1.46 (1.8) | 0.58 (0.12–2.25) | < 0.001a |
Abbreviations: ALP, alkaline phosphatase; AGR, albumin-to-globulin ratio; CAR, C-reactive protein-to-albumin ratio; CI, confidence interval; COPD, chronic obstructive pulmonary disease; DBil, direct bilirubin; DOHS, duration of hospital stay; GGT, gamma glutamyl transferase; hs-CRP, high-sensitivity C-reactive protein; IQR, interquartile range; LDH, lactate dehydrogenase; SD, standard deviation; SGOT, serum glutamate oxaloacetate transaminase; SGPT, serum glutamate pyruvate transaminase; SpO2, percentage saturation of oxygen; TBil, total bilirubin; TCS, total clinical severity score; T. protein, total protein.
a Denotes significance at p < 0.05 for independent t-test.
b Denotes significance at p < 0.05 for independent t-test for Mann–Whitney U test; The full names are mentioned in the abbreviation section.
The serum inflammatory parameters were significantly raised in the nonsurvivor group (p < 0.001). Renal profile parameters were also considerably increased in them (p < 0.001). Similarly, serum bilirubin and liver enzyme markers except for SGOT/SGPT were substantially elevated in the deceased patients. On the contrary, total serum protein (p = 0.002), albumin (p = 0.008), and AGR (p < 0.001) were significantly reduced. The CAR values were higher in the deceased group than the other (p < 0.001).
ROC and Cutoff Values of Predictor Variables for Mortality
The ROC, cutoff values, and its curves have been illustrated in ►Table 3 and ►Fig. 3. The sensitivity and specificity of hs-CRP at 59.5 mg/L were nearly 70% for nonsurvival. Similarly, serum LDH at 676.5 U/L showed a sensitivity of 70.1% and specificity of 74%, whereas serum ferritin at 550 ng/mL was close to 75% for mortality. Urea levels greater than 52.5 mg/dL depicted 85% specificity for mortality. Both sensitivity and specificity were almost identical for serum albumin of 3.23 g/dL (∼76%). Sensitivity was 70.1% for both AGR and CAR with 76.4% and 72.8% specificity, respectively. Serum values of parameters like direct bilirubin (0.19 mg/dL), SGOT (43.5 U/L), alkaline phosphatase (85.5 U/L), and total protein (6.6 g/dL) reported a sensitivity and specificity of more than 65%.

- Receiver operating characteristics (ROC) curves for laboratory variables to predict survival outcome. ALP, alkaline phosphatase; AGR, albumin-to-globulin ratio; CAR, C-reactive protein-to-albumin ratio; hs-CRP, high-sensitivity C-reactive protein; LDH, lactate dehydrogenase; SGOT, serum glutamate oxaloacetate transaminase.
| Laboratory variables (units) | AUC | SE | p-Value | Cutoff value | Sensitivity | 1-Sensitivity |
|---|---|---|---|---|---|---|
| hs-CRP (mg/L) | 0.765 | 0.02 | < 0.001a | 59.5 | 70.1 | 31.5 |
| LDH (U/L) | 0.779 | 0.021 | < 0.001a | 676.5 | 70.1 | 26.0 |
| Ferritin (ng/mL) | 0.817 | 0.019 | < 0.001a | 550 | 74.8 | 24.2 |
| 636.5 | 70.1 | 20.2 | ||||
| Urea (mg/dL) | 0.866 | 0.016 | < 0.001a | 52.5 | 70.9 | 15.3 |
| T. Protein (gm/dL) | 0.723 | 0.026 | < 0.001a | 6.6 | 66.9 | 0.32 |
| Albumin (gm/dL) | 0.847 | 0.016 | < 0.001a | 3.27 | 80.3 | 24.1 |
| 3.23 | 75.6 | 22.2 | ||||
| 3.17 | 71.7 | 20.1 | ||||
| AGR | 0.787 | 0.021 | < 0.001a | 0.97 | 70.1 | 23.6 |
| CAR | 0.794 | 0.019 | < 0.001a | 2.08 | 70.1 | 27.2 |
Abbreviations: AGR, albumin-to-globulin ratio; AUC, area under curve; CAR, C-reactive protein-to-albumin ratio; hs-CRP, high-sensitivity C-reactive protein; LDH, lactate dehydrogenase; SE, standard error; T. Protein, total protein.
a Denotes significance at p-value of 0.05.
Multiple Cox Regression and Kaplan–Meier Curves to Predict the Hazard Ratio for Mortality
Multiple cox regression and Kaplan–Meier curves to predict the HR for mortality during the hospital stay in the study population are illustrated in ►Table 4 and ►Fig. 4. The univariate analysis depicted a significantly high HR for nonsurvival patients with hyperkalemia (HR: 95% CI: 2.419; 1.96–2.99). The HR was 2% times more as age increased (p = 0.001). High SpO2 at admission reported a significant protective effect on mortality (HR: 95% CI: 0.931; 0.922–0.94). The risk for death was high by 0.065 and 0.086, with a unit increase in comorbidity and TCS score (p > 0.05). The risk for nonsurvival was increased with elevated serum levels of urea by 1.1%, creatinine by 15.6%, uric acid by 21.7%, sodium by 8.5%, and chloride by 6.1% (p < 0.001). Similarly, bilirubin and all liver enzymes but gamma glutamyl transferase reported significantly higher HR. The negative coefficient affirms the significant protective effect of high total serum protein, albumin, and AGR (p < 0.001). An increase in serum albumin would reduce the chances of death by a factor of 0.254 (95% CI: 0.196–0.33). A rise in CAR values increased the probability of death by 31.9% (p < 0.001). The chances for nonsurvival were 1.24 times with increasing serum globulin levels (p = 0.089).

- Kaplan–Meier curves of biochemical markers for survival function. ALP, alkaline phosphatase; AGR, albumin-to-globulin ratio; CAR, C-reactive protein-to-albumin ratio; CI, confidence interval; DBil, direct bilirubin; GGT, gamma glutamyl transferase; hs-CRP, high-sensitivity C-reactive protein; LDH, lactate dehydrogenase; SGOT, serum glutamate oxaloacetate transaminase; SGPT, serum glutamate pyruvate transaminase; SpO2, percentage saturation of oxygen; TBil, total bilirubin; TCS, total clinical severity score; TP, total protein. * denotes significance at p < 0.05.
| Variables | B Coefficient | p-Value | HR | 95% CI |
|---|---|---|---|---|
| Age (y) | 0.02 | 0.001a | 1.02 | 1.008–1.003 |
| SpO2 (%) | –0.071 | < 0.001a | 0.931 | 0.922–0.94 |
| Comorbidity score | 0.065 | 0.386 | 1.068 | 0.921–1.238 |
| TCS score | 0.086 | 0.145 | 1.09 | 0.97–1.22 |
| hs-CRP (mg/L) | 0.01 | < 0.001a | 1.01 | 1.007–1.012 |
| LDH(U/L) | 0.002 | < 0.001a | 1.002 | 1.002–1.003 |
| Ferritin (ng/mL) | 0.001 | < 0.001a | 1.001 | 1.001–1.002 |
| Urea (mg/dL) | 0.011 | < 0.001a | 1.011 | 1.009–1.013 |
| Creatinine (mg/dL) | 0.145 | < 0.001a | 1.156 | 1.099–1.215 |
| Uric acid (mg/dL) | 0.197 | < 0.001a | 1.217 | 1.159–1.278 |
| Na+ (mmol/L) | 0.081 | < 0.001a | 1.085 | 1.064–1.106 |
| K+ (mmol/L) | 0.884 | < 0.001a | 2.419 | 1.96–2.99 |
| Cl- (mmol/L) | 0.059 | < 0.001a | 1.061 | 1.035–1.09 |
| TBil (mg/dL) | 0.277 | < 0.001a | 1.319 | 1.201–1.45 |
| DBil (mg/dL) | 0.494 | < 0.001a | 1.639 | 1.41–1.903 |
| SGOT (U/L) | 0.002 | < 0.001a | 1.002 | 1.002–1.003 |
| SGPT (U/L) | 0.002 | < 0.001a | 1.002 | 1.001–1.003 |
| SGOT/SGPT | 0.217 | 0.023a | 1.243 | 1.031–1.498 |
| ALP (U/L) | 0.003 | < 0.001a | 1.003 | 1.002–1.005 |
| GGT (U/L) | 0.001 | 0.101 | 1.001 | 1–1.003 |
| T. Protein (gm/dL) | –0.601 | < 0.001a | 0.548 | 0.46–0.66 |
| Albumin (gm/dL) | –1.37 | < 0.001a | 0.254 | 0.196–0.33 |
| Globulin (gm/dL) | 0.217 | 0.089 | 1.242 | 0.97–1.593 |
| AGR | –2.948 | < 0.001a | 0.052 | 0.026–0.105 |
| CAR | 0.277 | < 0.001a | 1.319 | 1.246–1.397 |
Abbreviations: ALP, alkaline phosphatase; AGR, albumin-to-globulin ratio; CAR, C-reactive protein-to-albumin ratio; CI, confidence interval; COPD, chronic obstructive pulmonary disease; DBil, direct bilirubin; DOHS, duration of hospital stay; GGT, gamma glutamyl transferase; HR, hazard ratio; hs-CRP, high-sensitivity C-reactive protein; LDH, lactate dehydrogenase; SGOT, serum glutamate oxaloacetate transaminase; SGPT, serum glutamate pyruvate transaminase; SpO2, percentage saturation of oxygen; TBil, total bilirubin; TCS, total clinical severity score; T. protein, total protein.
a Denotes significance at p < 0.05; B denotes the coefficient for a variable when all other variables taken together; bold values denote significant for multivariate regression analysis.
The multivariate cox regression affirmed the association for SpO2 (p < 0.001), LDH (p = 0.006), ferritin (p = 0.004), hyperuricemia (p = 0.027), and hyperkalemia (p < 0.001), hypoalbuminemia (p = 0.002), and high CAR values (0.031) for nonsurvival, after adjusting for all the independent variables.
Discussion
Viewing the high death rate in this COVID-19 pandemic, early diagnosis of the infection becomes essential. Biomarkers like IL-6, ferritin, procalcitonin, D-dimer, and CK-MB are specified to know the disease severity. Only a few institutes and laboratories have the infrastructure for providing the testing facility. In addition, a limited number of trained personals are available to cater to a very high sample load that further limits the early reporting of the disease. By the time the patient has the laboratory report, the disease would have progressed to severity, enough to become challenging to revive. Hence, any laboratory bioparameter that would aid in monitoring the disease severity and diagnose the severity at an early condition would be quite a benefit. Therefore, routinely tested laboratory biomarkers that can be quickly investigated in all laboratories, easily accessible, and cheaper should be preferred for monitoring and diagnosing the severity of the disease condition at an early stage.
The study aimed to explore and provide insights regarding the influence of various demographic, clinical, and laboratory determinants on survival outcomes in patients admitted with COVID-19. The nonsurvivor group comprised 10.3% of the study population. The OR for death was 2.72 times for patients above 40 years of age. Males are more prone to death than females by a factor of 1.36. Nearly 46% of nonsurvivors died within 5 days of admission, with a median (IQR) of the duration of hospitalization was 6 (2–11) days. A high TCS score raised the risk for nonsurvival by 1.56. The serum inflammatory markers, renal parameters, liver enzymes, and CAR were significantly elevated in the deceased group. On the contrary, serum protein and albumin were reduced. Serum urea depicted highest specificity (85%) for nonsurvival at 52.5 mg/dL. Serum albumin (3.23 g/dL), AGR (0.97), and CAR (2.08) showed a sensitivity of more than 70% for mortality outcomes. After adjusting for all the predictor variables, SpO2 levels, serum LDH, ferritin, uric acid, potassium, albumin, and CAR values were affirmed as the potential biomarkers for mortality at the time of admission.
The mean (SD) age in severe cases was higher than the mild grade cases (►Table 2). It might be due to the coexistence of comorbidity in older age group subjects. Of the 127 nonsurvivors, 82 (64.6%) had associated comorbid conditions, and of them, nearly 63% were above 60 years (p < 0.001) (data not shown). In addition, weakened immunity in elderly persons further makes them susceptible to the severity of the disease.[3] A study by Li et al reported a median age of 59 (15–89) years.[18] The mean age of the deceased was 62.5 (13.7) years in Asirvatham et al study.[19] The median age in the present study (55 years) is in agreement with Li et al study.
A higher TCS score depicted a significant impact on mortality. Besides the co-existence of multiple comorbidities, clinical presentation at the time of admission greatly affects the recovery rate (►Fig. 1). Breathlessness was the main presenting feature in nearly 82% of nonsurvivors (►Fig. 2). It was followed by cough in 49.6% of them (►Fig. 2). On the contrary, 55.8 and 45% of survivors presented with cough and fever, respectively. Yang et al's study observed fever and cough as the predominant symptoms.[20] Leulseged et al study also reported cough (69.2%), dyspnea (34.5%), and fever (26.1%) in COVID-19 cases.[2] The study showed that presenting features such as a high respiratory rate due to dyspnea would increase the OR for mortality.
This study showed considerably elevated renal profile markers like urea, creatinine, and electrolytes in deceased cases. Hypertension was the most prevalent comorbid condition in the admitted patients, followed by DM (►Table 1). The presence of these comorbid conditions could have influenced the renal markers in the admitted patients. Hyperuricemia had confirmed a significant inverse relation with duration of hospital stay (data not shown). It also affirmed its potential for predictor biomarker for mortality (►Table 4). Hyperuricemia was also prevalent in COVID-19 deceased patients and correlated significantly with inflammatory cytokines, tumor necrosis factor-α, IL-6, and ferritin in 1,149 COVID-19-positive cases.[21] Collateral organ damage is one of the outcomes of SARS-CoV-2 infection. Inflammation and cytokine storm activated by the virus leads to systemic inflammation affecting the liver, kidney, heart, and other organs.[10,11] The patients usually succumb due to multiorgan failure. Uric acid, an end-product of purine metabolism, could be raised due to systemic organ damage and cell death.[21] It is characterized as a warning signal for systemic inflammation and host immune response.[22] The present study revealed that the probability of death was 2.68 times (95% CI: 1.13–6.39; p = 0.026) (data not shown). Serial monitoring of the renal profile, especially dyselectrolemia, is of utmost importance. These are the prime ions to be impaired due to reduced activity of angiotensin-converting enzyme-2 (ACE-2), the receptor for the SARS-CoV-2 virus.[12,13] Lippi et al study depicted significant hyponatremia in severe COVID-19 cases. Zimmer et al study reported hypernatremia in 6 out of 12 COVID-19 subjects admitted in an ICU.[13,23] Liu et al study documented higher OR for mortality (4.14) in patients with serum potassium greater than or equal to 5 mmol/L than those with lesser values.[24] In agreement with Liu et al study, hyperkalemia depicted a significant association with mortality (►Table 2). Liver damage revealed by high SGPT values and hypoalbuminemia correlated with severity. Hypoalbuminemia below 3.27 g/dL displayed a substantial increase in risk for death in COVID-19 cases. SGPT elevation signifies associated hepatic injury in the patients at admission.[25] The serum liver enzymes are frequently elevated in severe grade COVID-19 cases, but the clinical significance remains uncertain. It could be a marker for the systemic involvement of organs due to the cytokine storm. There is a low expression level of ACE-2 on hepatocytes that nullifies the direct effect of SARS-CoV-2 on liver damage.[26,27] However, serum albumin has shown a decreasing trend with progress in clinical severity. Hypoalbuminemia has been seen in more than 50% of COVID-19 hospitalized patients. It has been associated with severity and is considered as an independent predictor of mortality.[14,15]
Besides hypoalbuminemia, CAR also reported its potential for mortality predictor. Considering two biomarkers makes it a better biomarker as a mortality predictor. Values more than 2.08 were reported as a significant independent mortality predictor. The result agrees with Ranzani et al study that published CAR-associated significantly with mortality in patients admitted in an ICU.[28] Similarly, Karakoyun et al study too showed CAR as a significant marker for severity progression. CAR values of 0.9 depicted 70% sensitivity for severity in COVID-19 cases.[17] EL-Shabrawy et al study reported an HR of 26.5 for mortality after adjusting age and comorbidities.[16] Hs-CRP is one of the acute phase reactants that rises to any inflammation, and hypoalbuminemia is associated with severity. Hence, considering both markers would make it a better mortality predictor than considering the variables individually.
The study's greatest strength is its large sample size, providing an accurate statistical result for the predictor markers. Many studies usually focus on the inflammatory markers that are feasible to be tested in high-end instruments. The study has explored the basic parameters as predictors that can be easily estimated in every laboratory and help clinicians monitor the disease condition in a healthcare setting with limited facilities.
The study's primary limitation is its retrospective study design, and it is suggested that prospective serial monitoring of COVID-19 diagnosed patients would be more informative. Another limitation was that detailed comorbidity like disease duration, medication history, personal behavioral habits like smoking, alcohol intake could not be obtained from the record and could not be explored.
Conclusion
Early assessment of severity and immediate intervention is the key to the survival of COVID-19 patients. Monitoring specialized parameters is not feasible in remote areas and small-scale laboratories. Serial monitoring of serum biomarkers like uric acid, potassium, albumin, and CAR would help clinicians assess disease progression for severity in them for a survival outcome.
Abbreviations
| ACE-2 | Angiotensin-converting enzyme-2 |
| Alb | Albumin |
| ALP | Alkaline phosphatase |
| AGR | Albumin-to-Globulin ratio |
| ANOVA | Analysis of variance |
| ARDS | Acute respiratory distress syndrome |
| AUC | Area under curve |
| CAD | Coronary artery disease |
| CAR | C-reactive protein-to-Albumin ratio |
| CKD | Chronic kidney disease |
| CK-MB | Creatine kinase-MB |
| CKT | Creatine kinase total |
| Cl- | chloride ion |
| COPD | Chronic obstructive pulmonary disease |
| COVID-19 | Coronavirus disease of 2019 |
| CRP | C-reactive protein |
| CVA | Cerebro-vascular accident |
| DBil | Direct bilirubin |
| DM | Diabetes mellitus |
| GDM | Gestational Diabetes mellitus |
| GGT | Gamma glutamyl transferase |
| hs-CRP | high-sensitivity C-reactive protein |
| DOHS | Duration of Hospital Stay |
| HTN | Hypertension |
| HR | Hazard ratio |
| ICU | Intensive care unit |
| IL-6 | Interleukin-6 |
| IQR | Interquartile range |
| K+ | Potassium ion |
| LDH | Lactate dehydrogenase |
| LFT | Liver function test |
| Na+ | Sodium ion |
| OR | Odds ratio |
| RFT | Renal function test |
| ROC | Receiver operating characteristics |
| RT-PCR | Reverse transcriptase polymerase chain reaction |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus-2 |
| SCD | Sickle cell disease |
| SD | Standard deviation |
| SGOT | Serum Glutamate Oxaloacetate transaminase |
| SGPT | Serum Glutamate Pyruvate transaminase |
| SGOT/SGPT | Ratio of SGOT-to-SGPT |
| SpO2 | Percentage Saturation of oxygen |
| TB | Tuberculosis |
| TBil | Total bilirubin |
| TCS score | Total Clinical Severity score |
| TNF- α | Tumor necrosis factor- α |
| T. Protein | Total protein |
| WHO | World Health Organization |
Authors' Contribution
S. Patel, A.K. Behera designed the study and performed the statistical analysis. S. Patel and S. Shah wrote the protocol and the first draft of the manuscript. S. Prasad and G. Naik managed the data analyses of the study. R. Nanda and E. Mohapatra managed the literature searches and intellectual content. All authors read and approved the final manuscript.
Conflict of Interest
None declared.
Funding
None.
References
- Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) (2020) Accessed January 4, 2021 from: https://www.who.int/news/item/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov)
- [Google Scholar]
- Laboratory biomarkers of COVID-19 disease severity and outcome: findings from a developing country. PLoS One. 2021;16(03):e0246087.
- [CrossRef] [PubMed] [Google Scholar]
- COVID-19 and Your Health. People with certain medical conditions. Centers for Disease Control and Prevention 2021 Accessed January 4, 2021 from: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html
- [Google Scholar]
- Serum ferritin levels in inflammation: a retrospective comparative analysis between COVID-19 and emergency surgical non-COVID-19 patients. World J Emerg Surg. 2021;16(01):9.
- [CrossRef] [PubMed] [Google Scholar]
- Role of interleukin 6 as a predictive factor for a severe course of Covid-19: retrospective data analysis of patients from a long-term care facility during Covid-19 outbreak. BMC Infect Dis. 2021;21(01):308.
- [CrossRef] [PubMed] [Google Scholar]
- Meta-analysis of relation of creatine kinase-MB to risk of mortality in coronavirus disease 2019 patients. Am J Cardiol. 2020;130:163-165.
- [CrossRef] [PubMed] [Google Scholar]
- D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J Intensive Care. 2020;8(01):49.
- [CrossRef] [PubMed] [Google Scholar]
- Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan, China. Crit Care. 2020;24(01):525.
- [CrossRef] [PubMed] [Google Scholar]
- Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708.
- [CrossRef] [PubMed] [Google Scholar]
- Cytokine storm of COVID-19 and its impact on patients with and without chronic liver disease. J Clin Transl Hepatol. 2021;9(02):256-264.
- [CrossRef] [PubMed] [Google Scholar]
- COVID-19 and multiorgan failure: a narrative review on potential mechanisms. J Mol Histol. 2020;51(06):613-628.
- [CrossRef] [PubMed] [Google Scholar]
- Baseline electrolyte abnormalities would be related to poor prognosis in hospitalized coronavirus disease 2019 patients. New Microbes New Infect. 2020;37 100753
- [CrossRef] [PubMed] [Google Scholar]
- Electrolyte imbalances in patients with severe coronavirus disease 2019 (COVID-19) Ann Clin Biochem. 2020;57(03):262-265.
- [CrossRef] [PubMed] [Google Scholar]
- Hypoalbuminemia predicts the outcome of COVID-19 independent of age and co-morbidity. J Med Virol. 2020;92(10):2152-2158.
- [CrossRef] [PubMed] [Google Scholar]
- The association of low serum albumin level with severe COVID-19: a systematic review and meta-analysis. Crit Care. 2020;24(01):255.
- [CrossRef] [PubMed] [Google Scholar]
- Interleukin-6 and C-reactive protein/albumin ratio as predictors of COVID-19 severity and mortality. Egypt J Bronchol. 2021;15(01):1-7.
- [CrossRef] [Google Scholar]
- Diagnostic utility of C-reactive protein to albumin ratio as an early warning sign in hospitalized severe COVID-19 patients. Int Immunopharmacol. 2021;91 107285
- [CrossRef] [PubMed] [Google Scholar]
- Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199-1207.
- [Google Scholar]
- Who is dying from COVID-19 and when? An analysis of fatalities in Tamil Nadu, India. Clin Epidemiol Glob Health. 2021;9:275-279.
- [CrossRef] [PubMed] [Google Scholar]
- The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84 106504
- [CrossRef] [PubMed] [Google Scholar]
- Laboratory predictors of COVID-19 mortality: a retrospective analysis from Tongji Hospital in Wuhan. Mediators Inflamm. 2021;2021:6687412.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425(6957):516-521.
- [CrossRef] [PubMed] [Google Scholar]
- Hypernatremia-a manifestation of COVID-19: a case series. A A Pract. 2020;14(09):e01295.
- [CrossRef] [PubMed] [Google Scholar]
- Association between average plasma potassium levels and 30-day mortality during hospitalization in patients with COVID-19 in Wuhan, China. Int J Med Sci. 2021;18(03):736-743.
- [CrossRef] [PubMed] [Google Scholar]
- Liver impairment in COVID-19 patients: a retrospective analysis of 115 cases from a single centre in Wuhan city, China. Liver Int. 2020;40(09):2095-2103.
- [CrossRef] [PubMed] [Google Scholar]
- Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(02):631-637.
- [CrossRef] [PubMed] [Google Scholar]
- Update on new aspects of the renin-angiotensin system in liver disease: clinical implications and new therapeutic options. Clin Sci (Lond). 2012;123(04):225-239.
- [CrossRef] [PubMed] [Google Scholar]
- C-reactive protein/albumin ratio predicts 90-day mortality of septic patients. PLoS One. 2013;8(03):e59321.
- [CrossRef] [PubMed] [Google Scholar]






