Translate this page into:
Enterobacterial repetitive intergenic consensus-polymerase chain reaction analysis of a cluster of Elizabethkingia meningoseptica causing neonatal sepsis and meningitis
*Corresponding author: Ashoka Mahapatra, Department of Microbiology, All India Institute of Medical Sciences, Bhubaneswar, Odisha, India. meetasoka@yahoo.co.in
-
Received: ,
Accepted: ,
How to cite this article: Dey S, Mahapatra A, Sarathi S, Behera B, Sahoo T, Mishra B, Dey M. Enterobacterial repetitive intergenic consensus-polymerase chain reaction analysis of a cluster of Elizabethkingia meningoseptica causing neonatal sepsis and meningitis. J Lab Physicians. 2025;17:60-4. doi: 10.25259/JLP_32_2025
Abstract
Objectives
Elizabethkingia meningoseptica has emerged as a cause of healthcare-associated infections and outbreaks in intensive care units (ICUs) with high mortality because of its multidrug-resistant phenotype and adaptability to various environments. Strain typing is pivotal in detecting the cross-transmission of nosocomial pathogens and determining source tracing. Enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) typing has evolved as a simple, robust, cost-effective alternative to sequence-based typing methods for molecular epidemiological investigation of nosocomial outbreaks. The objectives of the study are to initiate an investigation of an outbreak after an alert of increased E. meningoseptica cases in neonatal ICU (NICU) (September 2021) and to conduct ERIC-PCR-based molecular epidemiological typing to assess clonal relatedness among the E. meningoseptica isolates of the outbreak.
Materials and Methods
E. meningoseptica isolates from blood and cerebrospinal fluid (CSF) were identified using standard microbiological techniques and final identification and antibacterial susceptibility by VITEK®2
Compact system (BioMérieux, France). Bacterial DNA was extracted using the spin column method (QIAprep Spin Miniprep kit-Qiagen). ERIC-PCR was performed following the procedure published in the literature.
Statistical analysis
Clonal relatedness among E. meningoseptica isolates was analyzed using ERIC-PCR fingerprints with the “Paleontological Statistics Software Package.”
Results
A cluster of 11 E. meningoseptica cases were encountered spanning over 15 months in NICU, and all were from both blood and CSF of 11 neonates, genetically related, pointing towards a common continuous source of the outbreak.
Conclusions
Heightened awareness among clinicians and microbiologists about E. meningoseptica, and stringent infection prevention and control measures are necessary to prevent such outbreaks in ICUs.
Keywords
Elizabethkingia meningoseptica
Enterobacterial repetitive intergenic consensus- polymerase chain reaction
Neonatal sepsis with meningitis
INTRODUCTION
Elizabethkingia meningoseptica, a non-fermenting Gram-negative bacillus, is ubiquitously distributed in nature and not a part of normal human flora. First identified as Flavobacterium meningosepticum by Elizabeth O’King in 1959 while investigating unclassified bacteria responsible for infant meningitis, the species underwent multiple taxonomic reclassifications. In 1994, it was renamed Chryseobacterium meningosepticum, and in 2005, following extensive phylogenetic studies, it was reclassified under the genus Elizabethkingia.[1]
In the past decade, E. meningoseptica has increasingly been isolated from hospital surroundings and clinical specimens, with its incidence rising from 6.8–13.1 to 26.6–39.9/100,000 admissions.[2] This organism has emerged as a significant healthcare-associated pathogen, contributing to alarmingly high morbidity and mortality.[2,3] Its resilience in chlorinated water, and disinfectant solutions, and its ability to colonize various hospital surfaces (e.g., sink basins, taps, saline bottles, and medical equipment such as respirators, intubation tubes, humidifiers, and nebulizers) highlight its potential as a cause of nosocomial infections.[1]
Moreover, E. meningoseptica poses several challenges to clinical microbiologists in accurate identification from closely related non-fermenters, the limited arsenal of effective antimicrobials, and the absence of established cut-offs for susceptibility determination.[2,3] E. meningoseptica clusters causing outbreaks are primarily described in neonatal intensive care units (NICU).[4] A recent study by Swami et al. described an outbreak of E. meningoseptica in a Level I NICU, leading to neonatal sepsis and meningitis over a 15-month period.[5] All clinically suspected cases were confirmed by laboratory identification of E. meningoseptica from blood and cerebrospinal fluid (CSF) cultures. These isolates were phenotypically related, showing similar antimicrobial susceptibility patterns. The outbreak was controlled by implementing stringent infection prevention and control (IPC) practices.[5]
Source attribution and determination of clonal relatedness remains the most significant challenges in tackling healthcare-associated clusters/outbreaks. Enterobacterial repetitive intergenic consensus (ERIC) sequences – 127 bp repetitive, imperfect palindromes – are promising tools for molecular typing, enabling the detection of clonal relatedness among bacterial isolates through PCR amplification of genomic regions between ERIC sequences and analysis of variation in these sequences.[6] Among various molecular typing methods available as epidemiologically important tools to detect cross-transmission of nosocomial pathogens, ERIC-PCR-based typing, in recent times, has evolved as a simple, rapid, and cost-effective alternative to sequence-based typing methods for molecular epidemiological investigation of healthcare-associated clusters and outbreaks.[2] In the present study, we intended to do molecular typing of the outbreak isolates of E. meningoseptica employing ERIC-PCR for assessing clonality.
MATERIALS AND METHODS
Identification of E. meningoseptica
Between September 2021 and November 2022, 22 isolates of E. meningoseptica were recovered from both blood and CSF samples of 11 neonates with sepsis and meningitis in the NICU. Most cases were observed in June and July 2022. Initial presumptive identification was based on colony morphology (non-lactose fermenting, translucent pale-yellow colonies on MacConkey agar), standard biochemical tests, resistance to colistin, and susceptibility to vancomycin and linezolid. Final identification and antimicrobial susceptibility were confirmed using the VITEK® 2 Compact system (BioMérieux, France).[7]
Molecular (ERIC-PCR) typing of the isolates
The 22 E. meningoseptica isolates (11 from blood and 11 from CSF) were subjected to ERIC-PCR to assess clonality. Genomic DNA was extracted from overnight cultures by spin column method using the QIAprep Spin Miniprep kit (Qiagen). The eluted DNA was quantified and checked for purity by measurement of sample absorbance at 260 nm and stored in microcentrifuge tubes at 20°C for further use.
ERIC-PCR was performed using the primers (F-5’-ATGTAAGCTCCTGGGGATTCAC-3’and R-5’-AAGTAAGTGACTGGGGTGAGCG-3’).[8] The procedure followed was as per published literature.[2,9] All reagents (Dream Taq™ Green PCR Master Mix and 50 ng of template DNA) were properly thawed and mixed to make the final reaction volume 25 µL. The thermocycler conditions included initial denaturation at 94°C for 3 min, 40 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 1 min, extension at 72°C for 2 min, followed by a final extension at 72°C for 10 min.
The amplicons were subjected to gel electrophoresis (1.5% agarose, 0.5 µg/mL ethidium bromide), and bands were visualized under ultraviolet light in an automated gel documentation system (Syngene, Germany). Clonal relatedness among the isolates of E. meningoseptica was analyzed using ERIC-PCR fingerprints with the “Paleontological Statistics Software Package.” Normalization steps were included to analyze the DNA polymorphism patterns of ERIC fingerprinting and ensure an adequate gelto-gel banding pattern. Band scoring was made by combining bands in each lane, and dendrograms were generated using Dice similarity co-efficient and interpreted using arithmetic averages with 1% optimization and 1% position tolerance. Isolates with a similarity exceeding 60% were considered to be clonally related.[9]
RESULTS
All the isolates demonstrated similar susceptibility and resistance patterns with 100% susceptibility to ciprofloxacin, levofloxacin, vancomycin, linezolid, and rifampicin and variable susceptibility to gentamicin (57.1%), amikacin (50%), piperacillin-tazobactam (28.6%), cefoperazone sulbactam (14.3%), cotrimoxazole (7.14%), and cefipime (7.14%).[2,3] Although the isolates in this study were susceptible to vancomycin, we observed suboptimal clinical outcomes following vancomycin monotherapy; hence, another sensitive antibiotic was added in all the cases, resulting in favorable outcomes with a median hospital stay of 6 days in all except for the death of two cases and complications of recurrent hydrocephalus in one case. Second blood culture after treatment could be obtained in eight cases, and they were sterile.
The fingerprints obtained from the ERIC typing of the 22 E. meningoseptica isolates (11 each from CSF and blood) consisted of 5–7 amplification bands ranging from 100 to 800 bp [Figure 1]. The dendrogram of the ERIC profiles identified 6 genotypes (A1-A6 in the same cluster) exhibiting >70% similarity [Figure 2].
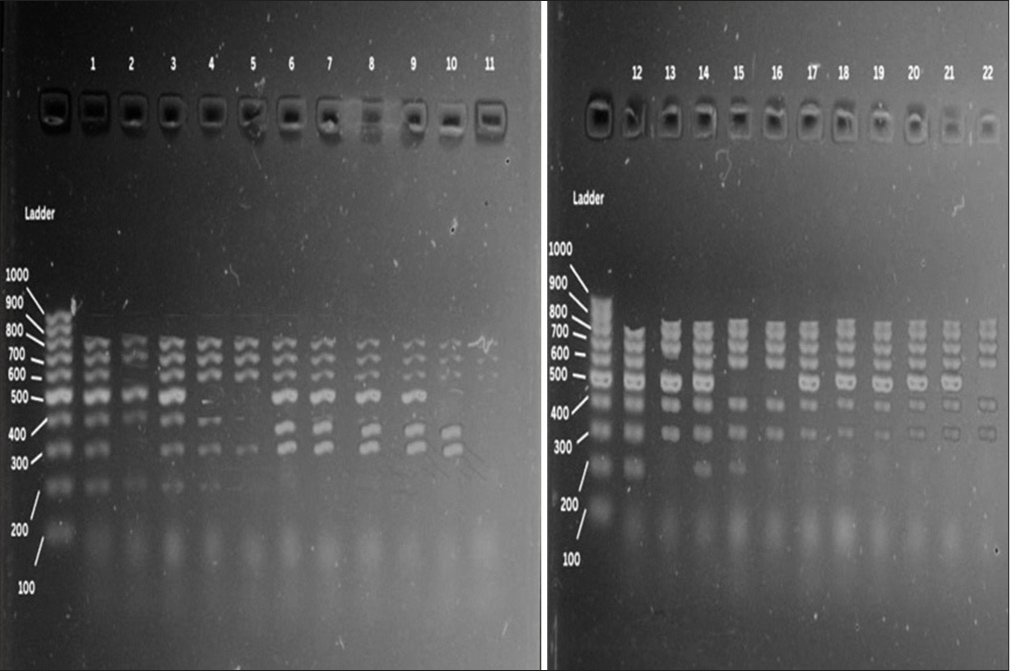
- DNA fingerprints of Elizabethkingia meningoseptica isolates generated by enterobacterial repetitive intergenic consensus polymerase chain reaction. Lanes 1–22 - clinical isolates, M: Molecular weight marker (100 bp).
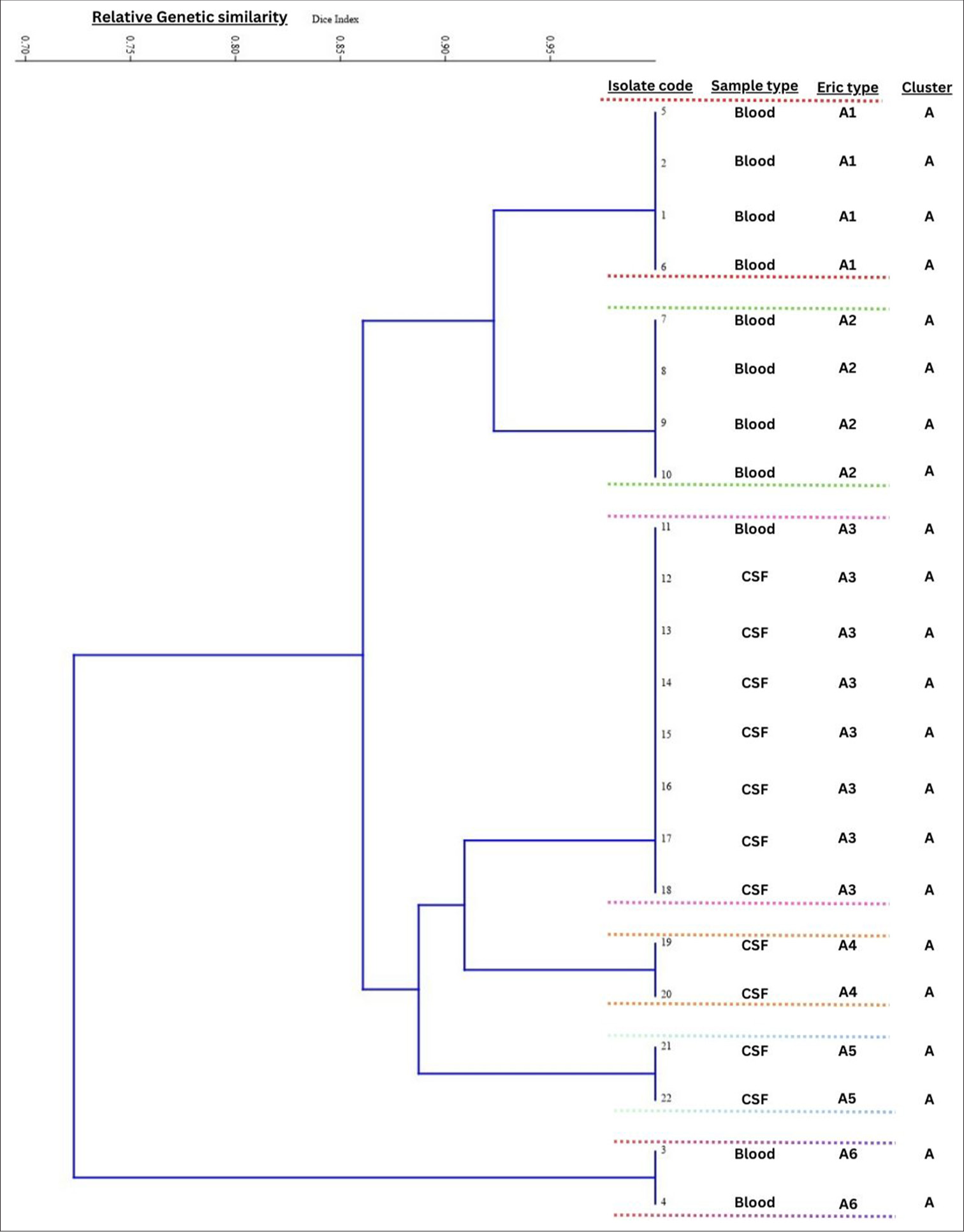
- Dendrogram of 22 Elizabethkingia meningoseptica isolates. The scale at the top of the dendrogram presents the relative genetic similarity. Enterobacterial repetitive intergenic consensus profiles identified 6 genotypes (A1-A6) exhibiting >70% similarity. CSF: Cerebrospinal fluid.
All the NICU doctors, nurses, and nursing mothers were trained for heightened awareness of IPC practices and strict hand hygiene compliance. Cohorting of cases, a dedicated nursing officer and equipment for each case, and strict implementation of contact precautions were followed. Disinfection practices of floors, surfaces, and equipment were reinforced with 70% alcohol and Cidex solution before and after every usage. Fumigation of two NICUs and cleaning and disinfection of water tanks were done.
Attempts were made to trace the source of the E. meningoseptica outbreak. In-use test of Reverse osmosis (RO) water for humidifier and pre-flush water in NICU-A grew Pseudomonas aeruginosa at 25°C and 37°C and post-flush water in NICU-B grew Aeromonas hydrophilia. The rest of the samples did not show growth of any microorganism. E. meningoseptica could not be traced from any environmental sample, even after repeated collection and testing. .
DISCUSSION
Healthcare-associated infections due to E. meningoseptica have increased in incidence among hospitalized patients in recent years, particularly in neonates, where it most commonly manifests as meningitis.[1,10] This denotes the neurovirulence of this organism in the neonatal population. Despite the knowledge of it being an emerging nosocomial pathogen, E. meningoseptica outbreaks are grossly under-reported. The possible reason is the unavailability of automated platforms for correctly identifying E. meningoseptica in resource-limited settings.[11] In our study, presumptive identification was based on colony morphology, Gram stain, oxidase testing, and antibiotic susceptibility, and was confirmed using the VITEK® 2 Compact system. Tai et al. and Kumar et al. also used the VITEK® 2 system for identification whereas Matrix-Assisted Laser Desorption Ionization–Time of Flight (MALDI-TOF) Mass Spectrometry had been used in a few other studies.[3,12-14]
During the clustering of E. meningoseptica cases from the NICU, we also had sporadic isolations of E. meningoseptica from other patient care areas. In previous reports, clusters of infection associated with E. meningoseptica are linked to water sources, sink drains, taps, respiratory equipment, tube feedings, syringes, flush solutions for arterial catheters, and antiseptic solutions.[2] A study from Taiwan reported isolation of E. meningoseptica from pacifier boxes, while another study from India reported isolation from breast pumps used in NICU.[3,12] However, our study could not ascertain the source despite extensive environmental culture-based surveillance involving water sources, disinfectants, and high-touch surfaces. Hence, we tried to determine clonality to determine if multiple clones are involved or if a single clone has clonal expansion. The clustering of all isolates in a single ERIC group, together with the sequence of appearance of new symptomatic cases over the 15-month period of our study, suggested a continuous common source.
Various molecular typing methods are available for assessing the outbreak strains’ clonal relatedness and source tracing. Different studies have mentioned that pulsed-field gel electrophoresis, repetitive element palindromic polymerase chain reaction (rep-PCR), and genome sequencing are commonly used techniques to establish indistinguishability between clinical and environmental isolates of E. meningoseptica.[3,13,15] However, we used ERIC-PCR typing to establish genetic relationships among clinical isolates of E. meningoseptica. ERIC-PCR is a robust, discriminatory typing tool for clinical isolates of Enterobacterales, and it had previously been successfully utilized as a method of genotypic characterization of various non-lactose fermenting organisms such as Pseudomonas, Stenotrophomonas, and Acinetobacter spp.[16-18] The advantages of ERIC-PCR are its simplicity, convenience of application, low cost per isolate, and minimal requirement for specialist laboratory equipment.
Genomic studies on E. meningoseptica are limited. Hu et al., in a recent study in 2023, performed a comprehensive whole genomic analysis of 47 E. meningoseptica isolates of six countries; some transnational isolates harbored identical single nucleotide polymorphisms (SNPs), suggesting super spreader potential of some clones. The study also concluded the severe outbreak-causing potential of E. meningoseptica within a hospital, as most of the outbreak-causing isolates from a single hospital harbored <10 SNPs.[19]
In published studies, E. meningoseptica outbreaks are mostly linked to water sources, including humidifier reservoirs. In the absence of source delineation, as in our case, it is recommended to enhance infection control practices, including isolation or cohorting of the infected neonates, dedicated nursing officer and equipment, strict implementation of contact precautions, and removal of all tap water sources from patient care areas along with frequent microbiological surveillance of hospital water supplies. Switching to sterile water for respiratory therapy devices in highly vulnerable areas, including neonatal ICU, could also be helpful.
CONCLUSIONS
It is time for health officials to heighten awareness about E. meningoseptica as an emerging pathogen among hospitalized patients. Despite being a single-center study with a limited sample size, our study can provide a baseline for future studies aiming to sequence many isolates in a multicentric design to facilitate the exchange of genetic data of this notorious pathogen with huge outbreak potential.
Author contribution
SD: Literature search, data collection, data analysis, interpretation, manuscript preparation; AM: Concept/design, literature search, data analysis, data interpretation, manuscript preparation, final approval; SS: Literature search, data analysis, data interpretation, manuscript preparation; BB: Concept/design, literature search, data analysis, data interpretation, manuscript preparation, final approval; MS: Literature search, data collection, data interpretation, manuscript preparation; BM: Literature search, data analysis, manuscript preparation, final approval; MD: Literature search, data analysis, data interpretation, manuscript preparation.
Ethical approval
The research/study was approved by the Institutional Review Board at AIIMS Bhubaneswar, Ref Number: IEC/AIIMS BBSR/PG Thesis/2020–21/59, dated 08th July 2020.
Declaration of patient consent
Patient consent was not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship: Nil.
References
- A Case series of Elizabethkingia meningosepticum Bacteremia in the cancer population. Cureus. 2021;13:e18627.
- [CrossRef] [Google Scholar]
- Elizabethkingia meningoseptica An important emerging pathogen causing healthcare-associated infections. J Hosp Infect. 2014;86:244-9.
- [CrossRef] [PubMed] [Google Scholar]
- Outbreak of Elizabethkingia meningoseptica sepsis with meningitis in a well-baby nursery. J Hosp Infect. 2017;96:168-71.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology and characteristics of Elizabethkingia spp. infections in Southeast Asia. Microorganisms. 2022;10:882.
- [CrossRef] [PubMed] [Google Scholar]
- Elizabethkingia meningoseptica outbreak in NICU: An observational study on a debilitating neuroinfection in neonates. Pediatr Infect Dis J. 2024;43:63-8.
- [CrossRef] [PubMed] [Google Scholar]
- Enterobacterial repetitive intergenic consensus (ERIC) Sequences in Escherichia coli Evolution and implications for ERIC-PCR. Mol Biol Evol. 2006;23:1156-68.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic diversity of various β-lactamase producing multidrug resistant Escherichia coli isolates from a tertiary care hospital using ERIC-PCR. Indian J Med Res. 2017;146:S23-9.
- [CrossRef] [PubMed] [Google Scholar]
- Enterobacterial repetitive intergenic consensus (ERIC) PCR based genetic diversity of Xanthomonas spp. and its relation to xanthan production. Iran J Microbiol. 2015;7:38-44.
- [Google Scholar]
- Microbiological characterization and clinical facets of Elizabethkingia bloodstream infections in a tertiary care hospital of Eastern India. Infect Drug Resist. 2023;16:3257-67.
- [CrossRef] [PubMed] [Google Scholar]
- Elizabethkingia in children: A comprehensive review of symptomatic cases reported from 1944 to 2017. Clin Infect Dis. 2018;67:144-9.
- [CrossRef] [PubMed] [Google Scholar]
- Successful control of Elizabethkingia meningoseptica outbreak in a neuro-surgical intensive care unit of a tertiary care hospital in North India. Int J Infect Control. 2018;14:1-6.
- [Google Scholar]
- Neonatal sepsis and meningitis caused by Elizabethkingia. Indian J Pediatr. 2021;88:598.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characteristics and antimicrobial susceptibility profiles of Elizabethkingia clinical isolates in Shanghai, China. Infect Drug Resist. 2020;13:247-56.
- [CrossRef] [PubMed] [Google Scholar]
- Application of whole genome sequencing to query a potential outbreak of Elizabethkingia anophelis in Ontario, Canada. Access Microbiol. 2019;1:e000017.
- [CrossRef] [PubMed] [Google Scholar]
- Bad design, bad practices, bad bugs: frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J Hosp Infect. 2013;85:134-40.
- [CrossRef] [PubMed] [Google Scholar]
- Biotyping of isolates of Pseudomonas aeruginosa isolated from human infections by RAPD and ERIC-PCR. Heliyon. 2021;7:e07967.
- [CrossRef] [PubMed] [Google Scholar]
- An epidemiological analysis of Stenotrophomonas maltophilia strains in a university hospital. Jpn J Infect Dis. 2004;57:37-40.
- [CrossRef] [PubMed] [Google Scholar]
- ERIC-PCR genotyping of Acinetobacter baumannii isolated from different clinical specimens. Saudi J Med Med Sci. 2018;6:13.
- [CrossRef] [PubMed] [Google Scholar]
- Probability of outbreaks and cross-border dissemination of the emerging pathogen: A genomic survey of Elizabethkingia meningoseptica. Microbiol Spectr. 2023;11:e01602-23.
- [CrossRef] [PubMed] [Google Scholar]






