Translate this page into:
Evaluation of simplified carbapenem inactivation method (sCIM) as a phenotypic method for rapid detection of carbapenemase-producing Enterobacterales: Study from a tertiary care hospital in North India
*Corresponding author: Prathyusha Kokkayil, Department of Microbiology, All India Institute of Medical Sciences, Patna, Bihar, India. drprathyushak@aiimspatna.org
-
Received: ,
Accepted: ,
How to cite this article: Archana A, Kokkayil P, DebBarma P, Priyadarshi K, Thakuria B. Evaluation of simplified carbapenem inactivation method (sCIM) as a phenotypic method for rapid detection of carbapenemase-producing Enterobacterales: Study from a tertiary care hospital in North India. J Lab Physicians. 2024;16:536-42. doi: 10.25259/JLP_102_2024
Abstract
Objectives:
This study aims to assess the feasibility of using the simplified carbapenem inactivation method (sCIM) for detecting carbapenemase production, specifically comparing its efficacy with the modified carbapenem inactivation method (mCIM), emphasizing methods applicable in low-resource settings and a minimal learning curve.
Materials and Methods:
To evaluate the performance of sCIM, 102 clinical isolates of carbapenem-resistant Enterobacterales (CREs) (detected by Kirby–Bauer disk diffusion technique) were selected, which had previously undergone both mCIM testing and genotyping detection of common carbapenemase-encoding genes. Polymerase chain reaction analysis of the isolates was done with specific primers targeting carbapenemase genes (blaNDM, blaIMP, blaVIM, blaSPM, blaKPC, and blaOXA48).
Statistical analysis:
Data were analyzed using the Statistical Package for the Social Sciences v.23. Quantitative variables were described using mean ± standard deviation or median (interquartile range). Categorical variables were described using proportions.
Results:
The sensitivity of sCIM was 90.43%, with a specificity of 87.5% when compared to the mCIM. The organism-wise analysis revealed notable sensitivity rates for Escherichia coli (93.44%) and Klebsiella pneumoniae (84.38%). Highlighting the efficacy of sCIM in identifying carbapenemase production in them. The specificity for sCIM remained high across all species, with 100% specificity for E. coli and 80% specificity for K. pneumoniae.
Conclusions:
The findings of our study support the efficacy of sCIM as a sensitive and specific method for the rapid detection of carbapenemase-producing CRE. The simplification of procedures and improved detection capabilities make sCIM a promising tool for timely and accurate identification, offering advantages over the traditional mCIM.
Keywords
Simplified carbapenem inactivation method
Phenotypic method
Carbapenemase-producing Enterobacterales
Polymerase chain reaction
INTRODUCTION
Gram-negative bacilli (GNB) such as Escherichia coli and Klebsiella pneumoniae belonging to Enterobacterales are among the common pathogens causing severe illness.[1] Various studies have shown a rapid emergence of carbapenem-resistant organisms in the recent decade.[2,3] Carbapenem-resistant Enterobacterales (CREs), as defined by the Centers for Disease Control and Prevention, encompass any member of the Enterobacterales family that exhibits resistance to imipenem, meropenem, doripenem, or ertapenem.[4] In addition to this resistance, CRE demonstrates insensitivity to other antibiotics within the beta-lactam category and also to other classes of drugs, such as aminoglycosides and fluoroquinolones, thereby constraining therapeutic options to polymyxins, tigecycline, and innovative beta-lactam-beta-lactamase inhibitor combinations.[5,6] This resistance poses substantial challenges in the management of CRE-associated infections, spanning bloodstream infections, pneumonia, urinary tract infections, and skin and soft tissue infections with a limited therapeutic arsenal.[7]
The associated infections show alarmingly high mortality rates of up to 50%.[8] The World Health Organization (WHO) in 2017 included CREs under the critical category in the global priority pathogens list.[9] The global prevalence of CRE has steadily risen, notably surging from 1% in 2013 to 43% in 2020 in North America, with varying rates reported in different regions of India.[8,10] Thus, there are limited antibiotic options, increased morbidity, mortality, and financial burdens.[11]
Carbapenem resistance within GNB can be attributed to multiple factors, including the production of carbapenemase enzymes, porin loss, enhanced efflux pump activity, and, infrequently, receptor mutation. The mechanisms underlying CREs predominantly revolve around the synthesis of carbapenemase enzymes, which are categorized into distinct groups exhibiting varying rates of dissemination and susceptibility profiles toward novel beta-lactam-beta-lactamase inhibitor combinations.[12] Many of the genes conferring carbapenem resistance are plasmid-mediated and have the potential for rapid dissemination across GN species.[2]
Identifying carbapenemase producers is crucial due to its significant role in carbapenem resistance within Enterobacterales. The Clinical and Laboratory Standards Institute (CLSI) endorses the modified carbapenem inactivation method (mCIM) for this purpose, given its high sensitivity (100%) and specificity (100%).[13,14] The mCIM, a modified protocol of the original carbapenem inactivation method (CIM) reported by van der Zwaluw et al. and Pierce et al., involves a broth incubation time of 4 h.[15,16] Recently, Jing et al. introduced a simplified CIM (sCIM) that directly applies test strains onto imipenem disks, omitting the broth incubation step and resulting in a more streamlined testing process and a slight reduction in costs.[17] However, the efficacy of sCIM for detecting Carbapenemase production has not been sufficiently evaluated. Therefore, this study aims to assess the feasibility of using sCIM for detecting Carbapenemase production by addressing the following questions: How does the performance of sCIM compare with mCIM in detecting carbapenemase production? What is the sensitivity and specificity of sCIM in identifying carbapenemase production across different species of CREs, with an emphasis on methods applicable in low-resource settings and a minimal learning curve?
MATERIALS AND METHODS
This cross-sectional study was done at the microbiology laboratory of a tertiary care hospital in Bihar, Eastern India. It was conducted between March 2023 and February 2024. A total of 102 clinical isolates of Enterobacterales which were resistant to any one of the carbapenems tested (namely meropenem or imipenem) by Kirby–Bauer disk diffusion method and interpreted according to the clinical breakpoints provided by CLSI M-100 document.[14] Different organisms isolated in a clinical specimen from the same patient and the same organism from different clinical specimens of the same patient were kept in the inclusion criteria. However, the same organism isolated from a particular specimen of the patient in the subsequent 7 days was excluded. The isolates were identified using standard microbiological and biochemical methods. All the 102 CRE isolates were subjected to mCIM and sCIM testing for carbapenemase production. Out of this, 49 isolates were further characterized by polymerase chain reaction (PCR) to detect common carbapenemase-encoding genes.[14]
The mCIM testing for the isolates was performed following the guidelines provided in document CLSI M100 34th edition.[14] A zone of inhibition of ≤15 mm, 16–18 mm, and ≥19 mm around the meropenem disk was interpreted as positive, indeterminate, and negative for carbapenemase, respectively.
The sCIM procedure was performed as described by Jing et al.[17] It involved the seeding of the Mueller–Hinton Agar (MHA) plate with 0.5 McFarland standard suspension of E. coli, ATCC 25922, employing the direct colony suspension method in alignment with routine disk diffusion protocols. Subsequently, 1–3 overnight cultured colonies of the test organisms on blood agar were uniformly smeared onto a 10 μg Imipenem disk (Hi-Media). This bacterial-coated disk was then positioned on an MHA plate previously inoculated with the indicator strain of E. coli, ATCC 25922. The antibiotic disc was placed so that the coated side came in contact with the indicator strain. Furthermore, plates were allowed to dry for at least 3–10 min before disc placement. A control setup, consisting of placing a plain 10 μg Imipenem disk (Hi-Media) without bacterial smearing on the MHA plate with the indicator strain, was included. The zone of inhibition was measured after 16–18 h incubation at 35 ± 2°C in ambient air [Figure 1].
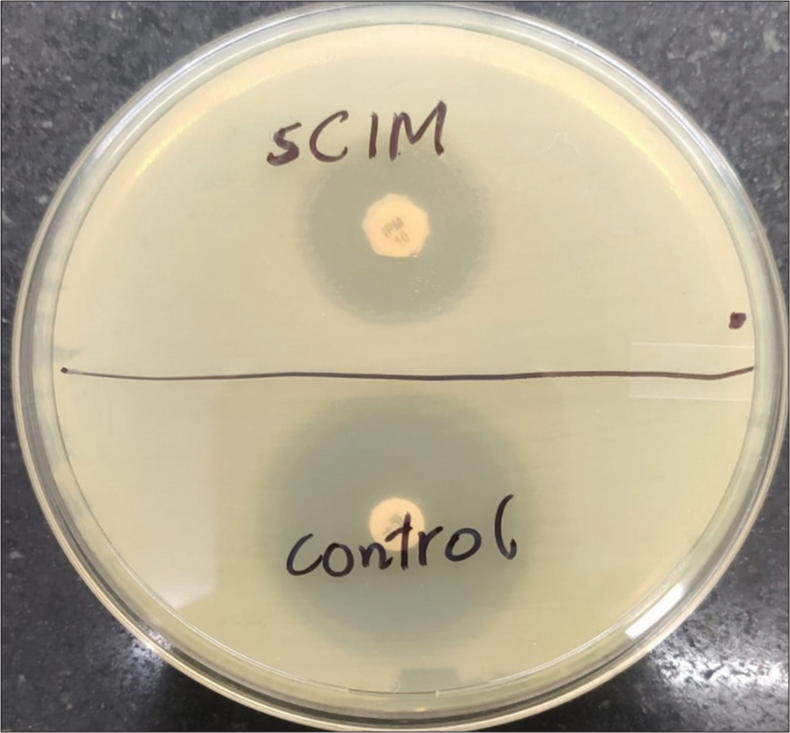
- Visualization of simplified carbapenem inactivation method testing of carbapenem-resistant Enterobacterales isolate: Imipenem disc (10 μg) with test strain smearing alongside the control disc with no bacterial smearing (for comparative analysis). sCIM: Simplified carbapenem inactivation method
Carbapenemase production was determined by evaluating the bacterial strain’s ability to hydrolyze imipenem, leading to unrestricted growth of the susceptible indicator strain. Carbapenemase production was indicated by a zone of inhibition with a diameter ranging from 6 to 20 mm or the presence of satellite growth of the colonies of E. coli ATCC 25922 with a zone diameter ≤22 mm. Conversely, a negative result was defined by a zone of inhibition ≥26 mm, whereas a result within the range of 23–25 mm was considered carbapenemase indeterminate.[17]
A total of 49 isolates underwent genotyping to assess the correlation between genotypic data and the performance of mCIM and sCIM in detecting carbapenemase production.
To genotype the test isolates, extraction of deoxyribonucleic acid (DNA) was done as per the protocol given by Ahmed and Dablool. Overnight bacterial growth of isolates on blood agar was used to prepare a cell suspension containing 107 cells/mL of isolates and underwent centrifugation at 4,500 rpm for 5 min at 4°C. Extraction of DNA was then performed using a modified boiling method, involving boiling the collected material at 100°C for 5 min with nuclease-free water. Thereafter, centrifugation at 3000 g for 10 min separated the upper aqueous phase containing DNA. After precipitating genomic DNA using ethanol, the resulting pellet was washed with cold 70% ethanol, dried, and then resuspended in aqueous TE buffer. Extracted DNA aliquots were stored at −20°C.[18]
Subsequently, PCR was conducted using specific primers targeting carbapenemase genes (blaNDM, blaIMP, blaVIM, blaSPM, blaKPC, and blaOXA48), and PCR conditions were set up according to a protocol outlined by Poirel et al. as shown in Table 1.[19] The PCR amplification was performed in a 25 μL reaction volume. Finally, the products of PCR were visualized through gel electrophoresis using a 2% agarose gel and an ultraviolet transilluminator.
| S. No. | Primer name | Sequence (5'-3') | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| 1. | blaVIM | Forward: GGTGTTTGGTCGCATATCGCAA Reverse: ATTCAGCCAGATCGGCATCGGC | 390 | Poirel et al.[19] |
| 2. | blaIMP | Forward: GGAATAGAGTGGCTTAAYTCTC Reverse: GGTTTAAYAAAACAACCACC |
232 | Poirel et al.[19] |
| 3. | blaNDM | Forward: GGTTTGGCGATCTGGTTTTC Reverse: CGGAATGGCTCATCACGATC |
621 | Poirel et al.[19] |
| 4. | blaKPC | Forward: CGTCTAGTTCTGCTGTCTTG Reverse: CTTGTCATCCTTGTTAGGCG |
798 | Poirel et al.[19] |
| 5. | blaOXA-48- like | Forward: GCGTGGTTAAGGATGAACAC Reverse: CATCAAGTTCAACCCAACCG | 438 | Poirel et al.[19] |
NDM: New Delhi metallo-beta-lactamase, OXA48: Oxacillinase, IMP: Imipenemase, KPC: Klebsiella pneumoniae carbapenemase, bp: base pairs, VIM: Verona integron-borne metallo-β-lactamase
The results obtained by sCIM were compared with mCIM and the gold-standard PCR detecting the carbapenemase-encoding genes, namely New Delhi metallo-beta-lactamase (NDM), oxacillinase (OXA-48), Verona integron-borne metallo-β-lactamase (VIM), imipenemase (IMP), Sao Paulo Metallo-β-lactamases (SPM), and Klebsiella pneumonia carbapenemase (KPC).
The data collected was entered and analyzed using the Statistical Package for the Social Sciences version 23. Quantitative variables were described using Mean ± standard deviation or Median (interquartile range). Categorical variables were described using proportions. A chi-square test was done to find the association between sCIM and mCIM.
RESULTS
Of the 102 CRE analyzed, 61.7% (63) were E. coli, 36.2% (37) were K. pneumonia, and 1.9% (2) were Citrobacter freundii. As the number of C freundiiwas low, they were included in the overall CRE analysis but not analyzed individually. The sensitivity of sCIM across all isolates was 90.43%, with a specificity of 87.5% when compared to the mCIM. Organism-wise analysis revealed notable sensitivity rates for E. coli (93.44%), highlighting the efficacy of sCIM in identifying carbapenemase production in them. K. pneumoniae exhibited a sensitivity of 84.38%, and specificity for sCIM remained high across all species, with 100% specificity for E. coli and 80% specificity for K pneumoniae A significant association was demonstrated between sCIM and mCIM using the Chi-square test (P < 0.0001). The overall performance metrics of the sCIM across all isolates have been summarized in Table 2.
| Organism | sCIM | mCIM | Total | |
|---|---|---|---|---|
| Positive | Negative | |||
| Escherichia coli | Positive | 57 | 0 | 57 |
| Negative | 4 | 2 | 6 | |
| Total | 61 | 2 | 63 | |
| Klebsiella pneumoniae | Positive | 27 | 1 | 28 |
| Negative | 5 | 4 | 9 | |
| Total | 32 | 5 | 37 | |
| Total | Positive | 85 | 1 | 86 |
| Negative | 9 | 7 | 16 | |
| Total | 94 | 8 | 102 | |
sCIM: Simplified carbapenem inactivation method, mCIM: Modified carbapenem inactivation method
The outcomes of the study assessing the performance of sCIM and mCIM in detecting carbapenemase production among 102 CRE isolates have been presented in Table 3 and Figure 2. The overall concordance was 92%, with species-specific variations.E. coli demonstrated a high agreement of 93.7%, accompanied by 4 very major errors (6.56%). K. pneumonia exhibited an 83.8% concordance, with 5 very major errors (15.63%) and 1 major error (20%).
| Organism | Count | Categorical agreement | Categorical disagreement | Major error |
Susceptible (mCIM negative) | Major error % | Very major error | Resistant (mCIM positive) | Very major error % |
|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | 63 | 59 | 4 | 0 | 2 | 0.00 | 4 | 61 | 6.56 |
| Klebsiella pneumoniae | 37 | 31 | 6 | 1 | 5 | 20.00 | 5 | 32 | 15.63 |
| Total | 102 | 92 | 10 | 1 | 8 | 12.50 | 9 | 94 | 9.57 |
sCIM: Simplified carbapenem inactivation method, mCIM: Modified carbapenem inactivation method, %: Percentage
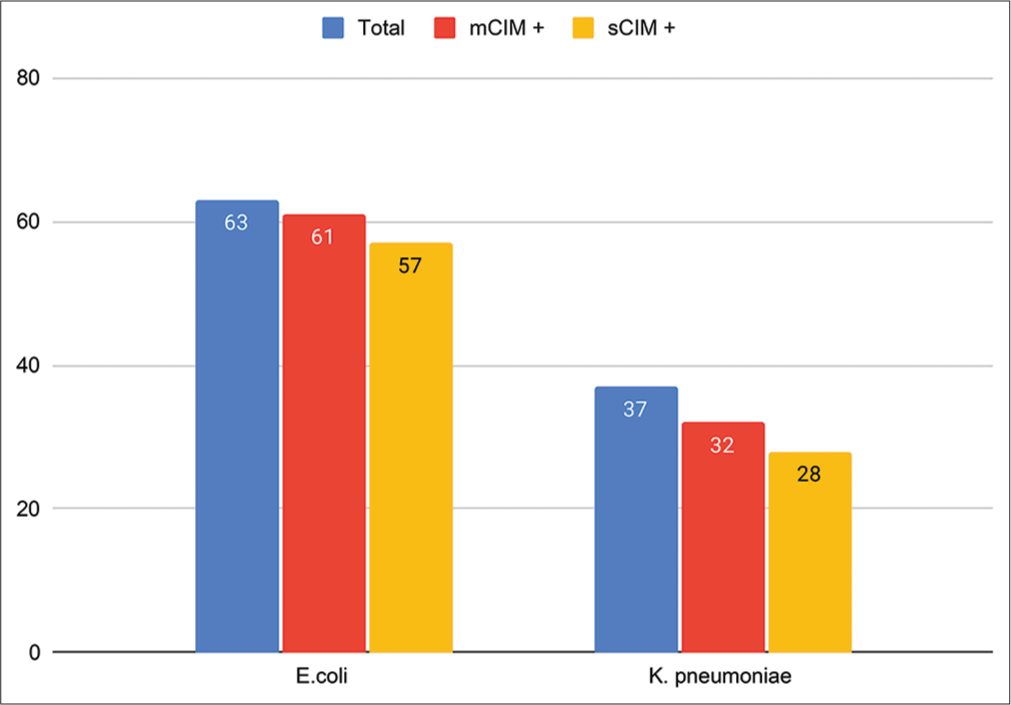
- Bar diagram showing comparison of modified carbapenem inactivation method and simplified carbapenem inactivation method results for the detection of carbapenemase-producing Enterobacterales (Escherichia coli and Klebsiella pneumoniae). sCIM: Simplified carbapenem inactivation method, mCIM: Modified carbapenem inactivation method
The details of the molecular characterization of isolates have been provided in Table 4. Genotyping data revealed diverse carbapenemase gene compositions. Among the 2 mCIM-positive but sCIM-negative E. coli isolates, three distinct carbapenemase genes were identified, including 2 NDM and 1 OXA-48 gene. Specifically, one isolate exclusively harbored the NDM gene, while the other exhibited a combination of OXA-48 and NDM genes.
| Total Escherichia coli | excl. NDM | NDM+OXA48 | excl. OXA48 | OXA48+IMP | excl. IMP |
|---|---|---|---|---|---|
| 28 | 10 | 4 | 12 | 1 | 1 |
| mCIM | 10 | 4 | 12 | 1 | 1 |
| mCIM % | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| sCIM | 9 | 3 | 12 | 1 | 1 |
| sCIM % | 90.00 | 75.00 | 100.00 | 100.00 | 100.00 |
| Total Klebsiella pneumoniae | excl. NDM | NDM+OXA48 | NDM+OXA48+KPC | NDM+OXA48+VIM | excl. OXA48 |
| 15 | 2 | 7 | 1 | 2 | 3 |
| mCIM | 2 | 7 | 1 | 2 | 3 |
| mCIM % | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| sCIM | 1 | 6 | 1 | 1 | 3 |
| sCIM % | 50.00 | 85.71 | 100.00 | 50.00 | 100.00 |
sCIM: Simplified carbapenem inactivation method, mCIM: Modified carbapenem inactivation method, %: Percentage, excl.: Exclusive, NDM: New Delhi metallo-beta-lactamase, OXA48: Oxacillinase, IMP: Imipenemase, KPC: Klebsiella pneumoniae carbapenemase, VIM: Verona integron-borne metallo-β-lactamase
The three K. pneumoniae isolates, which displayed very major errors, were found to harbor a total of 6 genes (3 NDM, 2 OXA-48, and 1 VIM). Molecular profiling showed distinct carbapenemase gene compositions within this subset: one isolate exclusively carried the NDM gene, another displayed a combination of NDM and OXA-48 genes, and the third exhibited a combination of NDM, OXA-48, and VIM genes.
Notably, the correlation between genotyping data and the ability of sCIM and mCIM to detect carbapenemase revealed varying efficacy, with sCIM detecting carbapenemase in 81% of NDM-harboring isolates, 90.32% of OXA-48-harboring isolates, and 100% of KPC and IMP-harboring isolates. In contrast, mCIM successfully detected carbapenemase in all isolates harboring carbapenemase genes, as demonstrated in Table 5.
| Genes | Count | mCIM | sCIM |
|---|---|---|---|
| excl. NDM | 10 | 10 | 9 |
| NDM+OXA48 | 4 | 4 | 3 |
| excl. OXA48 | 12 | 12 | 12 |
| OXA48+IMP | 1 | 1 | 1 |
| excl. IMP | 1 | 1 | 1 |
| Total | 28 | 28 | 26 |
sCIM: Simplified carbapenem inactivation method, mCIM: Modified carbapenem inactivation method, excl.: Exclusive, NDM: New Delhi metallo-beta-lactamase, OXA48: Oxacillinase, IMP: Imipenemase
DISCUSSION
Carbapenemase-producing CREs (CP-CREs) pose a significant clinical challenge and an undisputed threat to public health due to their resistance not only to carbapenems but also to various other classes of antimicrobial agents, rendering infections caused by these isolates particularly difficult to manage.[4] The mobility of carbapenemase genes, primarily located on plasmids, facilitates their transfer across different strains and species.[5] Given the clinical and epidemiological implications of infections caused by CREs, including CP-CRE, the rapid and accurate detection of CP-CRE is crucial for guiding appropriate therapeutic interventions and mitigating their spread within hospital settings.
Molecular assays have questionable use in a routine laboratory setting due to cost, availability, expertise, and feasibility. A rapid phenotypic test that holds the advantage for use in a high throughput laboratory with minimal cost is urgently required.
This study evaluated the sCIM as a potential diagnostic tool for detecting CP-CRE. Our findings revealed that sCIM demonstrated excellent sensitivity and specificity compared to the mCIM, with a sensitivity of 90.43% and specificity of 87.5%. In a study done by Hosoda et al., the sensitivity and specificity of sCIM were found out to be much lower i.e., 54.9% (28/51) and 84.2% (16/19) respectively, along with 10 indeterminate isolates when the interpretative criteria having zone of inhibition of ≤ 20 mm, 21–25 mm, and ≥ 26 mm were interpreted as positive, indeterminate, and negative, respectively.[20] Notably, the organism-specific analysis highlighted high sensitivity rates for E. coli (93.44%) and C. freundii (100%), indicating the effectiveness of sCIM in identifying carbapenemase production in these species. Although K. pneumoniae exhibited slightly lower sensitivity at 84.38%, the specificity of sCIM remained consistently high across all species, reaching 100% for E. coli and C. freundii and 80% for K. pneumoniae. These results align with a study by Jing et al., where sCIM showed 100% overall concordance with mCIM among 196 Enterobacterales, except for one result, which was false positive attributed to blaCTX-M in K. pneumoniae instead of a carbapenemase.[17] The sensitivity of this study also aligns with the findings of Yamada et al. (sensitivity and specificity of the sCIM were 97.0% and 100%).[21]
Despite its recognized utility, mCIM is not without limitations, such as the requirement for a broth incubation process followed by an overnight incubation period. However, its procedural simplicity contributes to a reduced risk of inter-technologist variability. sCIM was introduced as an alternative by Jing et al. with the primary aim of simplifying mCIM.[17] sCIM circumvents the 4-h broth incubation process, making it more adaptable to routine laboratory workflows. In addition, the study by Baeza et al. demonstrated the superior performance of sCIM in detecting metallo-beta-lactamases and its shorter time to results compared to mCIM.[22]
Limitation
All the 102 isolates did not undergo genotyping
Other isolates representative of Enterobacterales were not included because they were either not isolated during the study period or failed to show carbapenem resistance in vitro.
CONCLUSIONS
The findings of our study support the efficacy of sCIM as a sensitive and specific method for the rapid detection of CP-CRE. The simplification of procedures and improved detection capabilities make sCIM a promising tool for timely and accurate identification, offering advantages over the traditional mCIM. These findings have significant implications for clinical practice, emphasizing the potential role of sCIM in enhancing the diagnosis and management of CP-CRE infections in healthcare settings.
Author contributions
AA: Concept, design, definition of intellectual content, data acquisition, manuscript editing; PD: Literature search, data analysis, manuscript preparation; KP: Data analysis; PK and BT: Manuscript review.
Ethical approval
The research/study was approved by the Institute Ethical Committee vide letter no. AIIMS/Pat/IRC/2020/PGTh/Jan21/25.
Declaration of patient consent
Patient’s consent is not required for the present study, as the study was conducted on clinical isolates and there are no involvement of patients in the study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Carbapenem-Resistant enterobacterales in individuals with and without health care risk factors-Emerging infections program, United States, 2012-2015. Am J Infect Control. 2023;51:70-7.
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenemase-producing Gram-negative bacteria: Current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol. 2015;10:407-25.
- [CrossRef] [PubMed] [Google Scholar]
- The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The impact and evolution of a global menace. J Infect Dis. 2017;215(Suppl 1):S28-36.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial stewardship in the hospital setting: A narrative review. Antibiotics (Basel). 2023;12:1557.
- [CrossRef] [PubMed] [Google Scholar]
- Rationale and evidence for the use of new beta-lactam/beta-lactamase inhibitor combinations and cefiderocol in critically ill patients. Ann Intensive Care. 2023;13:65.
- [CrossRef] [PubMed] [Google Scholar]
- Occurrence of bla genes encoding carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii from intensive care unit in a tertiary care hospital. J Lab Physicians. 2018;10:208-13.
- [CrossRef] [PubMed] [Google Scholar]
- Further increases in carbapenem-, amikacin-, and fluoroquinolone-resistant isolates of Acinetobacter spp. and P. aeruginosa in Korea: KONSAR study 2009. Yonsei Med J. 2011;52:793-802.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of carbapenem resistant Enterobacteriaceae from a tertiary care hospital in Mumbai, India. J Microbiol Infect Dis. 2013;3:207-10.
- [CrossRef] [Google Scholar]
- WHO Publishes List of Bacteria for Which Antibiotics Are Urgently Needed. 2017. Geneva, Switzerland: WHO; Available https://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en [Last accessed on 2024 Jun 08]
- [Google Scholar]
- Molecular characterization of carbapenem-resistant enterobacterales collected in the United States. Microb Drug Resist. 2022;28:389-97.
- [CrossRef] [PubMed] [Google Scholar]
- ESBL, MBL and Ampc β Lactamases Producing superbugs-havoc in the intensive care units of Punjab India. J Clin Diagn Res. 2013;7:70-3.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp Ther Med. 2018;15:1143-9.
- [CrossRef] [Google Scholar]
- Comparison of four low-cost carbapenemase detection tests and a proposal of an algorithm for early detection of carbapenemase-producing Enterobacteriaceae in resource-limited settings. PLoS One. 2021;16:e0245290.
- [CrossRef] [PubMed] [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing In: CLSI Supplement M100 (34th ed). United States: Clinical and Laboratory Standards Institute; 2024.
- [Google Scholar]
- The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One. 2015;10:e0123690.
- [CrossRef] [PubMed] [Google Scholar]
- Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among enterobacteriaceae. J Clin Microbiol. 2017;55:2321-33.
- [CrossRef] [PubMed] [Google Scholar]
- The simplified carbapenem inactivation method (sCIM) for simple and accurate detection of carbapenemase-producing gram-negative bacilli. Front Microbiol. 2018;9:2391.
- [CrossRef] [PubMed] [Google Scholar]
- Quality Improvement of the DNA extracted by boiling method in Gram negative bacteria. Int J Bioassays. 2017;6:5347-9.
- [CrossRef] [Google Scholar]
- Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119-23.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of sCIM and other phenotypic detection methods for carbapenemase-producing enterobacterales. Microbiol Spectr. 2021;9:e0160821.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of the simplified carbapenem inactivation method as a phenotypic detection method for carbapenemase-producing enterobacterales. J Microbiol Methods. 2021;187:106273.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of five methods for detection of carbapenemases in Enterobacterales with proposal of a new algorithm. Clin Microbiol Infect. 2019;25:1286.e9-15.
- [CrossRef] [PubMed] [Google Scholar]






