Translate this page into:
Microbial Patterns and Antibiotic Susceptibility in Blood Culture Isolates of Septicemia Suspected Children in the Pediatrics Ward of a Tertiary Care Hospital
Address for correspondence: Muhammad Sarfraz Khan, MBBS Student, Rawalpindi Medical University, Rawalpindi, 46000, Pakistan (e-mail: dr.msk098@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objectives
This study aims to determine microbial patterns and antibiotic susceptibility to alert clinicians to the emerging pathogens that may pose a threat to the community, especially children.
Materials and Methods
This retrospective cross-sectional study was conducted in the Department of Pathology, Holy Family Hospital, Rawalpindi, Pakistan, from July 2019 to December 2019. Two samples were taken from two different sites of each patient at the time of fever (> 100° F) to avoid chances of contamination. A blood sample of 5 to 10 mL was drawn from each site. In a 50 mL brain heart infusion broth (BHIB), 5 to 10% blood was injected in two different bottles and incubated for 48 hours at 37°C. After 48 hours, subculture was done by streaking the drops of blood samples taken from BHIB on blood and MacConkey agar. Cultures were incubated in aerobic conditions at 37°C for 24 to 48 hours. For testing antibiotic susceptibility, criteria defined by the Clinical and Laboratory Standards Institute (CLSI) were followed. Microbes were identified under a microscope by observing their morphological characteristics after gram staining and applying biochemical tests. Antibiotic sensitivity test was performed using standard aseptic methods.
Statistical Analysis
Bacterial isolates and their susceptibility patterns were represented using frequencies and percentage charts.
Results
Out of 423 blood cultures, growth was recorded in 92 (21.75%) of the cultures with female to male ratio 2.1:1. The gram-positive bacteria accounted for 43.48% (n = 40), whereas gram-negative bacteria covered the majority 54.36% (n = 50). Among isolates, Staphylococcus aureus (42.39%) was the most common, followed by Acinetobacter spp. (17.39%) and Pseudomonas aeruginosa (14.13%). Acinetobacter spp. showed 0% susceptibility to amikacin and cefotaxime. All the isolates were 100% resistant to amoxicillin-clavulanic acid. S. aureus showed lower sensitivity for ceftazidime (0%), clindamycin (66.67%), ciprofloxacin (0%), clarithromycin (11.76%), and ceftriaxone (0%). Tigecycline showed 100% sensitivity for all isolates tested.
Conclusion
Gram-negative bacteria form the majority of isolates in our setup, with Acinetobacter as the most common species among them. The resistance against cephalosporins, penicillin, and fluoroquinolones shown by Acinetobacter, Pseudomonas, Salmonella, and Klebsiella is of grave concern. Among gram-positive bacteria, S. aureus has established resistance against multiple drugs. Limited and objective use of antibiotic therapy is a much-needed strategy under new guidelines.
Keywords
antibiotic susceptibility
blood culture
sepsis
pediatrics
Clinical and Laboratory Standards Institute (CLSI)
Introduction
Bacteria are responsible for many life-threatening infections by producing a wide range of virulence factors that enable them to overcome the human body's immune response and disseminate to remote organs and trigger a dysregulated host response.[1] Septicemia is a symptomatic extension of bacteremia throughout the body with clinical presentations that are representative of the infective pathogen and the organ system(s) involved.[2] The gold standard for the diagnosis of septicemia is a positive blood culture.[2]
Septicemia is considered as the leading common cause of death in United States and the most common cause of death among critically ill patients in noncoronary intensive care units.[3] Bloodstream bacterial infections have been shown to have mortality rate between 41.4 and 75.8%.[4] The incidence of bacteremia in Pakistan was reported as 912 (95% confidence interval [CI]: 648–1248) episodes per 1,00,000 child-years with a case fatality rate of 8%.[5] Among them, neonatal sepsis is a major cause of mortality in both developed and developing countries.[5] This might be due to delivery and postnatal follow-up, usually performed in a contaminated environment. This poses a higher chance of infection in neonates, which are already deficient in their inherent protective mechanisms, humoral and cellular immunity.[6] The changing pattern and frequent emergence of resistant bacteria make the problem worse. Children with septicemia present with fever, difficulty in breathing, tachycardia, malaise, inability to feed or lethargy, but those with asymptomatic bacteremia tend to show no obvious sign of illness.[2] Early detection and determination of antimicrobial susceptibility patterns have been shown to reduce the morbidity and mortality associated with bloodstream infections.[7] In developing countries, sepsis is common among children/neonates resulting from a combination of contaminated water, poor sanitation, indoor air pollution, crowding, low birth weight, insufficient immunization and nutrition, and increased need for invasive devices due to associated severe illnesses allowing pathogens to invade and multiply relatively unchecked in the body.[8] A paucity of robust data obtained through appropriately-powered clinical trials in children with a sepsis-related critical illness is one of the main challenges being faced with guiding the management of children with severe sepsis.[9,10]] This study aims to determine the early identification of bacteria and their patterns of susceptibility to antimicrobials to alert clinicians to the emerging pathogens that may pose a threat to the patients in the pediatric ward and serve as a guideline to the physician for empirical treatment with antibiotics of suspected bacteremia in children.
Materials and Methods
This descriptive cross-sectional study was conducted in the Department of Pathology, Holy Family Hospital, Rawalpindi, Pakistan, from July 2018 to December 2019. The study included all the consecutive blood cultures in children aged up to 13 years admitted to the Department of Pediatrics, Holy Family Hospital, Rawalpindi. The indication for blood cultures was clinical features adjudged by the attending clinician to be indicative of sepsis, especially fever without localized lesion. In total, 423 samples were selected, regardless of age and gender, using the simple convenient sampling technique. Two samples were taken from two different sites from each patient at the time of fever (> 100° F) to avoid chances of contamination. A blood sample of 5 to 10 mL was drawn from each site. In a 50 mL Brain heart infusion Broth (BHIB), 5 to 10% blood was injected in two different bottles and incubated for 48 hours at 37°C. After 48 hours, subculture was done by streaking the drops of blood samples taken from BHIB on blood and MacConkey agar. Cultures were incubated in aerobic conditions at 37°C for 24 to 48 hours. If no growth was noted, then the subculture was taken from the BHIB on 4th, 6th, and 7th day and incubated. The sample was labeled as no growth only if no culture was obtained even on 7th day. We used conventional blood culture system. Microbes were identified under a microscope by observing morphological characteristics after gram staining and applying biochemical tests. All patients had their blood cultures and sensitivities done at the time of admission in the ward. Antibiotic sensitivity pattern was done on Muller–Hinton agar using the Kirby Bauer disk diffusion method. All children under 13 years of age with signs and symptoms of bacteremia were included. Those who were terminally ill or had a history of taking antibiotics or both were excluded.
For testing antibiotic susceptibility, criteria defined by the Clinical and Laboratory Standards Institute (CLSI) was followed.[11] Antibiotic discs containing amikacin, amoxicillin-clavulanate (AMC), clindamycin (DA), ceftazidime (CAZ), clarithromycin (CLR), colistin (CT), cefotaxime (CTX), DA, cefoperazone+sulbactum (SCF), ciprofloxacin (CIP), ceftriaxone (CRO), cefepime (FEP), imipenem (IMI), tigecycline (TGC), vancomycin (VAN), penicillin (P), piperacillin/tazobactam (TZP), gentamicin (GN), TGC, trimethoprim-sulfamethoxazole (SXT), erythromycin (E), levofloxacin (LEV), meropenem (MEM), ampicillin (AMP), and moxifloxacin (MXF) were obtained and used as per the manufacturer's instructions. Vancomycin susceptibility for Staphylococcus aureus was determined using disc diffusion test. For that we used Mueller–Hinton agar and 30 µg discs of antibiotics discs. Organisms for which the vancomycin zone diameters are ≥ 15 mm were considered susceptible.
Information regarding age and sex was collected from notes made by doctors and nurses daily. Data were recorded from laboratory reports and analyzed using the Statistical Package for Social Sciences (SPSS) v.23.0 (IBM Inc.). Descriptive statistics were applied to find frequencies and percentages. Graph and tables were constructed.
Results
Microbial Patterns in Blood Culture Isolates Sections
Out of 423 blood cultures, growth was recorded in 92 (21.75%) of the cultures with female to male ratios of 2.1:1. Three hundred thirty-one (78.25%) accounted for negative cultures. The gram-positive bacteria accounted for 40 (43.48%), whereas gram-negative bacteria covered the majority 50 (54.35%). Candida spp. was found in 2 (2.17%) isolates. Highest number of bacterial isolates were S. aureus (39 [42.39%]) followed by Acinetobacter spp. (16 [17.39%]), Pseudomonas aeruginosa (13 [14.13%]), Salmonella typhi (11 [11.96%]) and Klebsiella pneumoniae (8 [8.7%]). S. aureus was the highest isolate among females (32 [82.05%]), while among males Sa. typhi (9 [81.82%]) dominated.
The species distribution of blood cultures is shown in ►Fig. 1
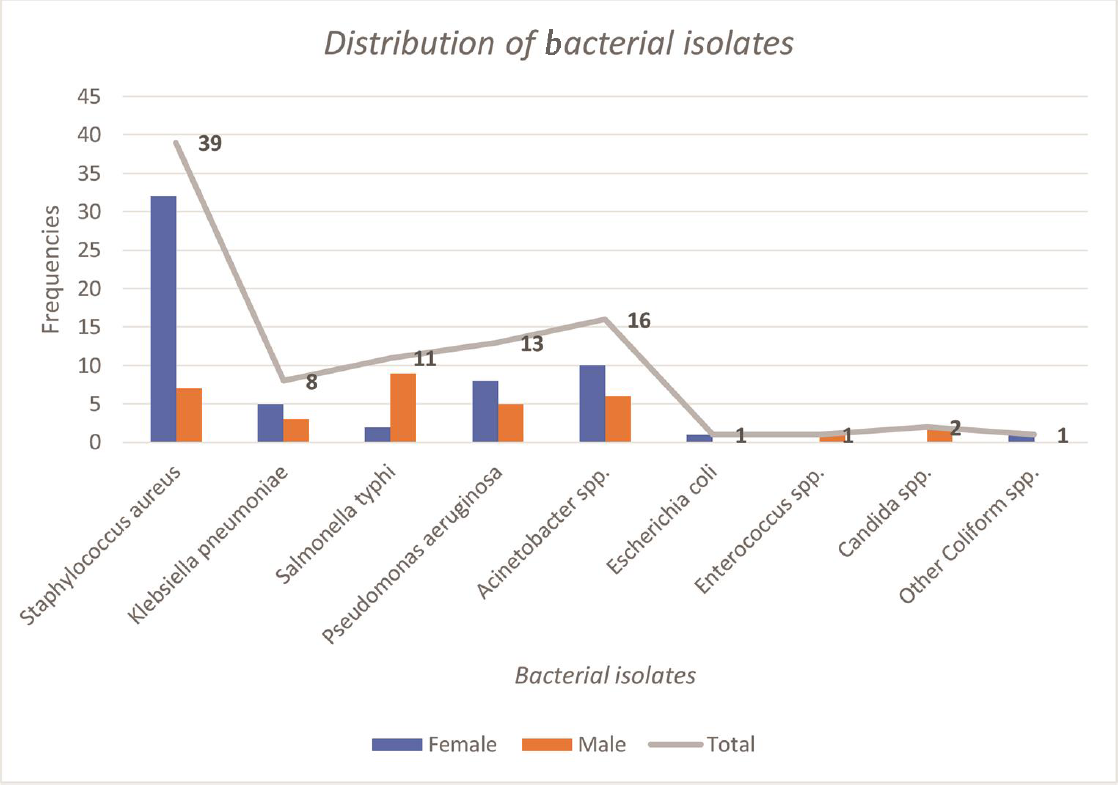
- Distribution of bacterial isolates in relation to sex among pediatric patients (n = 92).
Antibiotic Susceptibility in Blood Culture Isolates Sections
Among gram-positive bacteria, S. aureus showed lower sensitivity for ceftazidime (2.56%), DA (15.38%), CIP (7.69%), clarithromycin (43.58%), and CRO (2.56%). MXF showed variation in susceptibility toward S. aureus (82.05%) and Enterococcus spp. (100%). ►Table 1 shows antibiotic resistance patterns of gram-positive bacteria.
| Antimicrobial agents tested | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria isolated | Total | P | CIP | E | CLR | CAZ | GN | VAN | MXF | SXT | DA | AMP | CRO |
| n | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Staphylococcus aureus | 39 | 10 (25.64%) | 3 (7.69%) | 24 (61.53%) | 17 (43.58) | 1 (2.56%) | 29 (74.35%) | 24 (61.53%) | 32 (82.05%) | 14 (35.89) | 6 (15.38%) | 1 (2.56%) | 1 (2.56%) |
| Enterococcus | 1 | – | – | 1 (100%) | – | – | 0 | 0 | 0 | – | 1(100%) | 1 (100%) | – |
| Total | 40 | 10 (25%) | 3 (7.5%) | 25 (62.5%) | 17 (42.5%) | 1(2.5%) | 29 (72.5%) | 24(60%) | 32(80%) | 14 (35%) | 7(17.5%) | 2(5%) | 1(2.5%) |
Abbreviations: AMP, ampicillin; CAZ, ceftazidime; CIP, ciprofloxacin; CLR, clarithromycin; CRO, ceftriaxone; GN: gentamicin; DA, clindamycin; E, erythromycin; GN, gentamicin; MXF, moxifloxacin; P, penicillin; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
Note: 0=no resistance; – shows the antibiotics were not tested against this organism.
Gram-Negative Profile
Among gram-negative bacteria, Acinetobacter was the most resistant, showing 0% susceptibility to the majority of antibiotics. Pseudomonas aeruginosa was susceptible to most of the antibiotics. Sa. typhi, P. aeruginosa, and Escherichia coli showed high resistance towards AMC, CAZ, CRO, CTX, SCF, and FEP. TGC showed 100% sensitivity for the majority of isolates tested. ►Table 2 shows antibiotic resistance patterns of gram-negative bacteria.
| Antimicrobial agents tested | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria isolated | Total no. | MEM | CIP | CRO | CTX | CAZ | GN | IMI | SXT | SCF | AMC | TZP | FEP | MXF | TGC |
| n | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Escherichia Coli | 1 | – | – | 0 | – | 1 (100%) | – | 1 (100%) | – | – | 1 (100%) | 0 | 1 (100%) | – | 0 |
| Klebsiella spp. | 8 | 2 (25%) | 6 (75%) | 8 (100%) | 2 (25%) | 7 (87.5%) | 2 (25%) | 8 (100%) | – | 3 (37.5%) | 6 (75%) | 8 (100%) | 8 (100%) | 1 (12.5%) | |
| Acinetobacter spp. | 16 | 01 (6.25%) | 12 (75%) | 15 (93.75%) | 11 (68.75%) | 15 (93.75%) | 2 (12.5%) | 14 (87.5%) | 2 (12.5%) | 7 (43.75%) | 15 (93.75%) | 14 (87.5%) | 4 (25%) | 01 (6.25%) | 0 |
| Salmonella typhi | 11 | 0 | 7 (63.63%) | 10 (90.90%) | 4 (36.36%) | 11 (100%) | 4 (36.36%) | 9 (81.81%) | 4 (36.36%) | 3 (27.27%) | 11 (100%) | 0 | 8 (72.72%) | 2 (18.18%) | 0 |
| Pseudomonas aeruginosa | 13 | 0 | 13 (100%) | 10 (76.92%) | 4 (30.76%) | 11 (84.61%) | – | 11 (84.61%) | – | 8 (61.52%) | 1 (7.69%) | 12 (92.30%) | 10 (76.92%) | 0 | |
| Other coliforms spp. | 1 | – | – | 0 | – | 1 (100%) | – | 1 (100%) | – | 0 | 1 (100%) | 0 | 1 (100%) | – | 0 |
| Total | 50 | 3 (6%) | 38 (76%) | 43 (86%) | 21 (42%) | 46 (92%) | 8 (16%) | 44 (88%) | 6 (12%) | 21 (42%) | 35 (70%) | 34 (68%) | 32 (64%) | 4 (8%) | 0 |
Abbreviations: AMC, amoxicillin-clavulanate; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CTX, cefotaxime; FEP, cefepime; GN, gentamicin; IMI: imipenem; MEM, meropenem; MXF, moxifloxacin; SCF, cefoperazone+sulbactum; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; TZP, piperacillin/tazobactam.
Note: 0=no resistance; – shows the antibiotics were not tested against this organism.
Discussion
In this study, a paramount difference was recorded between males and females in the overall blood culture growth positive rate (females 67.3 vs. males 32.7%, p = 0.038). However, this finding disproves several other studies that reported high culture positivity in males compared with females.[12] The reason for this difference is unclear and requires further study with large sample size. Bacteria are increasingly becoming resistant to conventional antibiotics in clinical and nonclinical settings.[13] The incidence of bacteremia varies widely in the pediatric population. In our study, bacteremia's positivity came to be 21.75%, while a study in Nigeria showed the incidence of bacteremia to be 48.9% with S. aureus covering 48.7% of it.[3] Our study reported the incidence of gram-negative bacteremia to be higher (54.36%) than gram-positive (43.48%). In a similar study in India, gram-negative organisms were 88.8% of total isolates and gram-positive was 10.38%.[2]
Among gram-positive isolates, S. aureus was followed by Enterococcus (1.09%), unlike a study done by Marina Thomas where S. aureus was among the most frequent isolates (50.6%), but Enterococcus faecalis followed it as second most frequent gram-positive isolate (9.3%).[14] Our study S. aureus showed maximum resistance against penicillin and CIP, whereas zero resistance was shown against vancomycin and least against gentamycin, amikacin, and cefotaxime, this is in consistence with a study in Cambodia.[15] It was noted that in neonatal intensive care unit neonatal pneumonia was empirically treated with broad range antibiotics in all babies without culture and sensitivity, this injudicious use of antibiotics in turn accounts for such high rate of hospital acquired VRSA.
In our research, the most frequent isolates among gram-negatives were Acinetobacter species (17.39%), followed by P. aeruginosa (14.13%) and Sa. typhi (11.96%). This is in contrast with an Indian study, which reported gram-negative bacteremia with the most frequent isolate Klebsiella (47.1%) followed by Salmonella (16.2%), Pseudomonas (7.99%), and Acinetobacter (6.19%).[2]
Coming to Acinetobacter (17.39%), which is the most frequent isolate in our study among gram-negative bacteria, has become highly resistant to antibiotics over the years, requiring an aggressive treatment.[16] In our study, Acinetobacter showed high resistance (100%) to multiple drugs such as amikacin, amoxicillin-clavulanic acid, ceftazidime, CIP, IMI, and CRO. This is in concordance to another study where these drugs also showed maximum resistance to different strains of Acinetobacter.[15]
Pseudomonas aeruginosa is a motile gram-negative rod that belongs to the family Pseudomonadaceae, disseminated widely in nature, and it is highly prevalent in the hospital setting since it encourages bacterial growth.[17] In our research, it exhibited 14.13% prevalence and marked resistance to several drugs. In our study, it showed resistance to piperacillin/tazobactam (92.31%), LEV (100%), cefotaxime (100%), ceftazidime (90.9%), and FEP (90%). These results are in agreement with other studies.[18,19]
The third most common gram-negative bacillus in our study came to be Sa. typhi (11.96%). In a study conducted in Nigeria, the same organism isolated from blood samples of children reported being 0.7% of total isolates,[3] while a study in India showed 15.08% isolates to be Sa. typhi.[2] Our study reported 100% sensitivity to TGC against Sa. typhi. IMI showed 88.89% of sensitivity. Fifty percent of sensitivity was shown by amikacin, gentamycin, and azithromycin. Among fluoroquinolones, CIP showed 63.64%, and MXF showed 18.198% resistance. Hundred percent resistance was recorded by amoxicillin, SXT and cephalosporins. This is in concordance with an Indian study where cephalosporins showed maximum resistance.[2]
The other gram-negative bacilli of our study included E. coli and other coliforms (1.09% each). However, in a study done by Fox-Lewis et al, E. coli showed 47.2% of positive isolates[15] while a study in Pakistan showed that both E. coli and other coliforms were 5% of isolates each.[20] In our study, E. coli showed sensitivity to amikacin, while other coliforms were resistant to it. These organisms showed sensitivity to CIP, TGC, and cefoperazone/sulbactam (100%). Both of these organisms were highly resistant to IMI, clarithromycin, and cephalosporins tested. These findings are somehow similar to a study done by Madhu Sharma, where cephalosporins showed some degree of resistivity.[2] During the evaluation of possible bacterial pathogens, 2.17% of cultures yielded Candida species. Madhu Sharma recorded this percentage to be 0.79%.[3]
Worldwide emergence of resistant bacteria is a real threat to a favorable outcome to conventional forms of infection in the hospital settings and community. A careful selection of antibiotics is required to avoid treatment failure and the spread of resistant strains. Poor infection control practices with inappropriate and unjust use of antibiotics are the main driving force behind the tremendous increase in antimicrobial resistance strategies like combination therapy, antibiotic restriction, and antibiotic cycling must be put in use to prevent the emergence of resistance. Association of public health policy with active research on the determinants of the evolution of resistance to antimicrobial agents and its reversal is much-needed action. This study should be done on a larger scale involving different departments of a tertiary care hospital to describe antibiotic resistance trends in all age groups.
Conclusion
Gram-negative bacteria form the majority of isolates in our setup with Acinetobacter most common species among them. TGC and colistin were found to be effective against Acinetobacter. The resistance against cephalosporins, penicillin, and fluoroquinolones shown by Acinetobacter, Pseudomonas, Salmonella, and Klebsiella is of grave concern. Among gram-positive bacteria, S. aureus has established resistance against multiple drugs. However, TGC and vancomycin remain effective against this pathogen. Limited and objective use of antibiotic therapy is a much-needed strategy under new guidelines. Antibiotic stewardship should be promoted.
Consent to Participate
Informed consent was obtained from all individual participants included in the study. Personal details of subjects were kept under confidentiality.
Acknowledgments
None.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
This study is not supported by any kind of funds.
References
- Gram-positive and gram-negative bacterial toxins in sepsis: a brief review. Virulence. 2014;5(01):213-218.
- [CrossRef] [PubMed] [Google Scholar]
- Acute Care Handbook. Physical Therapists. Saunders 2013 Available at: Accessed Jan 6, 2020
- [Google Scholar]
- Epidemiology of severe sepsis. Virulence. 2014;5(01):4-11. DOI: 10.4161/viru.27372
- [CrossRef] [PubMed] [Google Scholar]
- Bacteremia is associated with excess long-term mortality: a 12-year population-based cohort study. J Infect. 2015;70(02):111-126.
- [CrossRef] [PubMed] [Google Scholar]
- Incidence of pneumonia, bacteremia, and invasive pneumococcal disease in Pakistani children. Trop Med Int Health. 2010;15(09):1029-1036.
- [CrossRef] [PubMed] [Google Scholar]
- Study of prevalence and antimicrobial susceptibility pattern of bacteria isolated from blood cultures. J Biol Sci. 2009;9:249-253.
- [CrossRef] [Google Scholar]
- Predictive model for bacterial late-onset neonatal sepsis in a tertiary care hospital in Thailand. BMC Infect Dis. 2020;20(01):151. DOI: 10.1186/s12879-020-4875-5
- [CrossRef] [PubMed] [Google Scholar]
- Clinical trials in pediatric sepsis: what's the (end) point? Pediatr Crit Care Med. 2017;18(03):296-297.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology and etiology of childhood pneumonia. Bull World Health Organ. 2008;86(05):408-416.
- [CrossRef] [PubMed] [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI Supplement M100 (28th). Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
- [Google Scholar]
- Novel approaches to the development of anti-sepsis drugs. Expert Opin Drug Discov. 2014;9(05):523-531.
- [CrossRef] [PubMed] [Google Scholar]
- Bacteremia among Jordanian children at Princess Rahmah Hospital: Pathogens and antimicrobial susceptibility patterns. Iran J Microbiol. 2010;2(01):22-26.
- [Google Scholar]
- Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(09):597-602.
- [Google Scholar]
- Antimicrobial resistance in invasive bacterial infections in hospitalized children, Cambodia, 2007-2016. Emerg Infect Dis. 2018;24(05):841-851.
- [CrossRef] [PubMed] [Google Scholar]
- Bacteriologic profile and antibiogram of blood culture isolates in a pediatric care unit. J Lab Physicians. 2010;2(02):85-88.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of pseudomonas aeruginosa and its antibiotic sensitivity pattern. J Nepal Health Res Counc. 2019;17(01):109-113.
- [CrossRef] [PubMed] [Google Scholar]
- Bacteriological profile and antibiogram of cultures isolated from tracheal secretions. Cureus. 2019;11(06):e4965. DOI: 10.7759/cureus.4965
- [CrossRef] [Google Scholar]
- Resistance pattern of Pseudomonas aeruginosa isolated from clinical specimens in a Tertiary Hospital in Northeastern Nigeria. Inter J Microbiol. 2009;8:1-6.
- [CrossRef] [Google Scholar]
- Antibiotic resistance pattern of Pseudomonas aeruginosa isolated from urine samples of Urinary Tract Infections patients in Karachi, Pakistan. Pak J Med Sci. 2015;31(02):341-345.
- [Google Scholar]





