Translate this page into:
Molecular Characterization of Extended Spectrum β-Lactamase Producing Escherichia coli and Klebsiella pneumoniae Isolates and Their Antibiotic Resistance Profile in Health Care-Associated Urinary Tract Infections in North India
Address for correspondence: Sheetal Verma, MBBS, MD, Department of Microbiology, King George's Medical University, Chowk, Lucknow, 226003, Uttar Pradesh, India (e-mail: dr.sheetal2001@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Healthcare-associated urinary tract infections (HAUTIs) caused by gram-negative pathogens have emerged as a global concern. So far, little is known about the epidemiology of extended-spectrum β lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in HAUTIs in India. The study was carried to determine the antibiotic resistance pattern and ESBL-producing genes in E. coli and K. pneumoniae strains isolated from HAUTIs in a tertiary institute in North India.
Methods
A total of 200 consecutive, nonduplicate clinical isolates of E. coli and 140 isolates of K. pneumoniae from hospitalized patients with UTI were collected during a period of 1 year. Strains were studied for the presence of ESBL genes (blaCTX-M1, blaCTX-M2, blaCTX-M9, blaCTX-M15, blaSHV, blaTEM, blaOXA-1, blaVEB, blaPER-2, and blaGES) by multiplex polymerase chain reaction using gene-specific primers.
Results
ESBL was detected in 82.5% (165 out of 200) isolates of E. coli and 74.3% (104 out of 140) isolates of K. pneumoniae by phenotypic confirmatory testing. From 269 phenotypically positive ESBL isolates, blaTEM (49.4%) was the most common genotype followed by blaCTX-M1 (31.97%), blaOXA-1 (30.1%), and blaSHV(11.9%) either alone or in combination. In the present study, blaCTX-M-15 (84.89%) was the most common blaCTX-M1-type ESBL. In total, 2.6 and 5.2% of the isolates were positive for PER-2 and VEB genes, respectively.
Conclusion
To the best of our knowledge, this is the first study on ESBL resistance patterns and ESBL-producing genes in HAUTIs in North India. Our study reports high occurrence with ESBL types CTX-M-1, CTX-M-15, TEM, and SHV. Minor ESBL variants OXA-1, VEB-type, and PER-2-type β-lactamase are also emerging in HAUTIs infections in North India.
Keywords
β-lactamase
drug resistance
ESBL
heathcare-associated urinary tract infections
PCR
Introduction
Health care-associated urinary tract infections (HAUTIs) are common and contribute to severe morbidity and high fatality rate in hospitalized patients. About 80% of HAUTIs occur in patients who undergo instrumentation of the urinary tract including catheter-associated urinary tract infections.[1] The emergence of extended-spectrum β-lactamase (ESBL) is frequently associated with prolonged hospital stay, increased treatment cost, and limited treatment options especially wide-spectrum antibiotics.[2]
ESBLs have been reported in India most frequently in Enterobacteriaceae and cause difficulty in the management of infections in intensive critical care settings. The prevalence of ESBLs resistance varies greatly in geographic areas, and the trend keeps on changing over time at a rapid pace. CTX-M, TEM, SHV-type ESBL have become prevalent worldwide including in India, and enzymes are becoming increasingly expressed by pathogenic bacterial strains in hospitals and have the potential for widespread dissemination.[3] ESBLs such as OXA-1, PER-type, GES-type, and VEB-type have also been reported from various countries. Different investigators across India have reported prevalence ranging from 7.0 to 91% by different methods to detect ESBL resistance.[4]
Polymerase chain reaction (PCR)-based molecular methods are rapid, accurate, and have better sensitivities for detecting ESBL-resistant genes than conventional phenotypic methods. They help clinicians with a targeted approach for the treatment and also help in containment of outbreaks and the implementation of infection control policies.[5] Most data including the distribution of the etiologic agents and resistance patterns of UTIs are community based, and there are limited studies concerning the HAUTIs in India.
This study was conducted to determine the occurrence of ESBL-encoding genes (CTX-M1, CTX-M2, CTX-M9, CTX-M15, TEM, SHV, OXA-1, VEB, PER-2, and GES type) in Escherichia coli and Klebsiella pneumoniae isolates recovered from HAUTIs and to study their pattern of antimicrobial resistance.
Material and Methods
The present prospective study was conducted among patients admitted to the intensive care unit (> 3 days after hospital admission) in a tertiary hospital in North India over a period of 1 year from June 2018 to May 2019. Recruited cases were patients with positive E. coli and K. pneumonaie urine culture (≥ 105 colony forming units (CFU)/mL for midstream and ≥ 104 for samples collected by catheter and any count of bacteria for samples acquired by suprapubic aspiration), and wet mount showed significant pyuria (> 10 white blood cells per high-power field).[4] Only those cases were included where only a single strain was obtained on culture using cystine–lactose–electrolyte-deficient agar. Isolates were identified by matrix-assisted laser desorption/ionization time of flight mass spectrometry (bioMérieux) at species level following the manufacturer's recommendations and stocked in 50% glycerol solution at −80°C for further analysis. This study was approved by the institutional ethical committee (83rd ECM IIA/P14).
Extended-Spectrum β Lactamase Screening by the Disk Diffusion Method
The isolates were screened by one of these drugs (30 μg of cefotaxime, ceftazidime, ceftriaxone, or aztreonam) on a standard disk diffusion procedure. The zones 27 mm or lesser for cefotaxime, 22 mm or lesser for ceftazidime, 25 mm or lesser for ceftriaxone, or 27 mm of lesser for aztreonam were considered as probable ESBL producers and were further confirmed by double disk diffusion testing.
Extended-Spectrum β Lactamase Confirmation by Double Disk Diffusion Testing
In brief, disks containing cefotaxime with and without clavulanic acid (10 μg) were placed 20 mm apart on the surface of a Muller Hinton agar (Hi-Media) plate previously inoculated with the test organism and incubated at 37°C for 18 hours. A test result was considered positive if there was an increase of 5 mm or higher in the zone diameter for the combination of antimicrobial agent and clavulanic acid compared with the antibiotic alone. The test was done as per recommended by the Clinical and Laboratory Standards Institute (CLSI) document M100-S21.[6]K. pneumoniae ATCC 700603 was used as the ESBL-positive control, and E. coli ATCC 25922 was used as the negative control.
Antibiotic Susceptibility Testing and Minimum Inhibitory Concentration Determination
The antibiotic susceptibility pattern of the ESBL-producing isolates to a panel of antibiotics was performed by the Kirby Bauer disk diffusion technique as per CLSI criteria. The isolates were tested against following antibiotics: norfloxacin (30 μg), nitrofurantoin (300 μg), trimethoprim and sulfamethoxazole (1.25/23.75 μg), amikacin (30 μg), gentamicin (10 μg), piperacillin-tazobactam (100/10 μg), cefepime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), cefuroxime (30 μg), cefepime (30 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg), ertapenem (10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), and tetracycline (30 μg). Colistin was also tested by the broth microdilution method. All disks and salt were obtained from Hi-Media, Mumbai, India. E. coli ATCC 25922 was used as quality control strain. The minimum inhibitory concentration (MIC) values (μg/mL) for cefotaxime, ceftazidime, and cefepime were determined using E-tests (Hi-Media, Mumbai, India) and interpreted according to manufacturer's instructions.
Analysis of ESBL Genes
Genotypic analysis for blaCTX-M1 group, blaCTX-M2 group, blaCTX-M9 group, blaCTX-M15, blaSHV, blaTEM, blaOXA-1, blaPER-2, blaVEB, and blaGES were performed on phenotypically confirmed ESBL isolates. The primers of the genes were used as described previously (►Table 1).[5,7,8] Detection of β-lactamase genes was performed using a set of three multiplex PCR assays and one monoplex PCR assay. Clinical strains carrying those β-lactamase genes were used as positive controls.
| Target | Primer sequence (5′→3′) | Amplicon (bp) | Reference |
|---|---|---|---|
| blaCTX-M1 group | F: 5′-TTAGGAAATGTGCCGCTGTA-3′ R: 5′-CGATATCGTTGGTGGTACCAT-3′ |
878 | [7] |
| blaCTX-M2 group | F: 5′-AAACAGAGCGAGAGCGATAAG-3′ R: 5′-GGGTAAAGTAGGTCACCAGAAC-3′ |
324 | [7] |
| blaCTX-M9 group | F: 5′-GCA GAT AAT ACG CAG GTG-3′ R: 5′-CGG CGT GGT GGT GTC TCT-3′ |
164 | [7] |
| blaCTX-M-15 | F: 5′-CACGTCAATGGGACGT-3′ R: 5′-GAAAGGCAATACCACCGGT-3′ |
811 | [This study] |
| blaTEM | F: 5′-CATTTCCGTGTCGCCCTTATTC-3′ R: 5′-CGTTCATCCATAGTTGCCTGAC-3′ |
800 | [5] |
| blaSHV | F: 5′-AGCCGCTTGAGCAAATTAAAC-3′ R: 5′-ATCCCGCAGATAAATCACCAC-3′ |
713 | [5] |
| blaOXA-1 | F: 5′-TCAACTTTCAAGATCGCA-3′ R: 5′-GTGTGTTTAGAATGGTGA-3′ |
609 | [5] |
| blaVEB | F: 5′-CATTTCCCGATGCAAAGCGT-3′ R: 5′-CGAAGTTTCTTTGGACTCTG-3′ |
648 | [8] |
| sblaPER-2 | F: 5′-GCTCCGATAATGAAAGCGT-3′ R: 5′-TTCGGCTTGACTCGGCTGA-3′ |
520 | [8] |
| blaGES | F: 5′-AGTCGGCTAGACCGGAAAG-3′ R: 5′-TTTGTCCGTGCTCAGGAT-3′ |
399 | [8] |
DNA Extraction and Polymerase Chain Reaction
Bacterial DNA isolation for all experiments was done by the boiling lysis method as previously described.[9] Five colonies of each test organism was grown in 1.5 mL of Tryptic Soy Broth (Himedia, Mumbai, India) at 37 °C for 18 to 24 hours, followed by centrifugation at 10,000 × g for 5 minutes. After discarding the supernatant, the pellet was resuspended in 300-μL nuclease-free water. The cells were then boiled for 5 minutes, followed by immediate incubation on ice for 2 minutes. Finally, the sample was centrifuged at 10,000 × g, and the supernatant was directly used for PCR. This crude DNA extract (2 μL) was used as a template for all the reactions. All PCR amplifications were performed using Thermal Cyclers (Applied Biosystems, Foster City, United States).
Multiplex Polymerase Chain Reaction-I for the Detection of blaCTX-M1, blaCTX-M2, and blaCTX-M-9 Groups
Reaction mixture was optimized in volume of 25 μL containing 2 μL of DNA template prepared from each isolate; Taq buffer (10 ×): 3 μL; dNTP mix (10 mM): 1 μL; MgCl2 (25 mM): 1.5 μL; three forward primers (10 pmol/μL): each 0.5 μL; three reverse primers (10 pmol/μL): each 0.5μL; TaqDNA polymerase (1 U/μL): 1 μL; and nuclease-free water: 13.5 μL. The amplification condition was initial denaturation for 10 minutes at 94°C, followed by 30 cycles of 94°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute, and final extension of 72°C for 10 minutes.
Multiplex Polymerase Chain Reaction-II for the Detection of blaTEM, blaSHV, and blaOXA-1 Genes
Reaction mixture was optimized in the volume of 25 μL containing 2 μL of DNA template prepared from each isolate; Taq buffer (10 ×): 3 μL; dNTP mix (10 mM): 0.5 μL; MgCl2 (25 mM):1 μL; three forward primers (10 pmol/μL): each 0.5 μL; three reverse primers (10 pmol/μL): each 0.5 μL; TaqDNA polymerase (1 U/μL): 1 μL; and nuclease-free water: 15.5 μL. The amplification condition was initial denaturation for 5 minutes at 94°C, followed by 30 cycles of 94°C for 30 seconds, annealing at 54°C for 30 seconds, extension at 72°C for 1 minute, and final extension of 72°C for 10 minutes.
Multiplex Polymerase Chain Reaction-III for the Detection of blaVEB, blaPER-2, and blaGES Genes
Reaction mixture was optimized in the volume of 25 μL containing 2 μL of DNA template prepared from each isolate; Taq buffer (10 ×): 2.75 μL; dNTP mix (10 mM): 0.5 μL; MgCl2 (25 mM): 1 μL; three forward primers (10 pmol/μL): each 0.75 μL; three reverse primers (10 pmol/μL): each 0.5 μL; TaqDNA polymerase (1 U/μL): 1 μL; and nuclease-free water: 15 μL. The amplification condition was initial denaturation for 10 minutes at 94°C, followed by 30 cycles of 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 30 seconds, and final extension of 72°C for 10 minutes.
Statistical Analysis
Descriptive statistics including frequencies were analyzed using Statistical Package for the Social Sciences (SPSS), version 20 (SPSS Inc. Chicago IL, United States). All categorical variables were expressed as numbers and proportions.
Results
Extended-Spectrum β Lactamase Screening and Confirmatory Testing
In the study, 200 nonrepetitive bacterial isolates of E. coli and 140 isolates of K. pneumoniae were recovered from hospitalized patients. In total, 88% (176 out of 200) isolates of E. coli and 83.6% (117 out of 140) isolates of K. pneumoniae showed resistance to at least one of the third generation cephalosporin by phenotypic screening tests. ESBL was detected among 82.5% (165 out of 200) isolates E. coli and 74.3% (104 out of 140) isolates of K. pneumoniae by confirmatory testing (►Table 2). The majority of patients were between the age group of 31 to 75 years in both groups (►Fig. 1). Age ranged from 18 to 90 years, with a mean age of 44.38 ± 20.39 years, and it was observed that age was not significantly associated with the incidence of both E. coli and K. pneumoniae infection (p > 0.05).
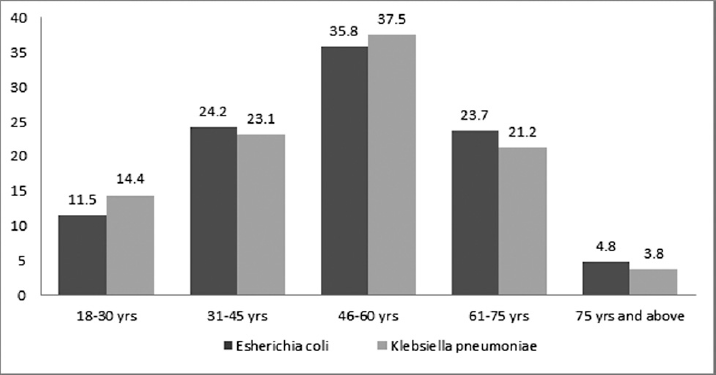
- Age-wise frequency distribution of ESBL producing isolates. ESBL, extended-spectrum β lactamase.
| Bacterial isolates | ESBL screening by the disk diffusion method | ESBL confirmation by double disk diffusion testing | |||
|---|---|---|---|---|---|
| Cefotaxime (30 μg) N (%) | Ceftazidime (30 μg) N (%) | Ceftriaxone (30 μg) N (%) | Aztreonam (30 μg) N (%) | Cefotaxime (30 μg) with clavulanic acid (10 μg) N (%) | |
| E. coli (200) | 122 (61) | 152 (76) | 157 (78.5) | 111 (55.5) | 165 (82.5) |
| K. pneumoniae (140) | 101 (72.1) | 104 (74.3) | 99 (70.7) | 76 (54.3) | 104 (74.3) |
| Total (340) | 223 (65.5) | 256 (75.3) | 256 (75.3) | 187 (55) | 269 (79.1) |
Abbreviations: ESBL, extended-spectrum β lactamase.
Antibiotic Susceptibility Patterns
The resistance of ESBL producers to various antibiotics in E. coli versus K. pneumoniae was as follows: norfloxacin (88.4 vs. 79.8%), ciprofloxacin (83 vs. 85.5%), meropenem (82.4 vs. 89.4%), ceftriaxone (80.6 vs. 78.8%), ceftazidime (76.9 vs. 79.8%), trimethoprim/sulfamethoxazole (72.7 vs. 78.8%), and imipenem (53.3 vs. 67.3%), respectively, thus revealing multidrug resistance. The susceptibility to colistin was 100 vs. 100%, followed by nitrofurantoin (84.3 vs. 22.1%), while amikacin was 75.2 vs. 33.7%, piperacillin and tazobactam 67.9 vs. 60.6%, ertapenem 65.5 vs. 41.4%, and gentamicin 64.3 vs. 47.2%. Overall, K. pneumoniae isolates were observed to have higher resistance rates than E. coli strains especially amikacin (66.3%), gentamicin (52.8%), nitrofurantoin (77.9%), cefepime (82.6%), and ertapenem (58.6%). There was higher resistance seen in ESBL-producing isolates compared with non-ESBL-producing isolates (►Table 3). MIC determination of the ESBL-positive isolates to third-generation cephalosporins like ceftriaxone, ceftazidime, and cefotaxime showed high resistance varying in the range of 32 to less than 256 μg/mL.
| Antimicrobial agents | ESBL-producing E. coli (N = 165) (%) | Non-ESBL-producing E. coli (N = 35) (%) | ESBL-producing K. pneumoniae (N = 104) (%) | Non-ESBL-producing K. pneumoniae (N = 36) (%) | Total ESBL-producing isolates (N = 269) (%) | Total non-ESBL-producing isolates (N = 71) (%) |
|---|---|---|---|---|---|---|
| Amikacin | 41 (24.8) | 13 (37.1) | 69 (66.3) | 12 (33.3) | 110 (40.9) | 25 (35.2) |
| Aztreonam | 103 (62.4) | 12 (34.2) | 66 (63.4) | 10 (27.8) | 169 (62.8) | 22 (30.9) |
| Ceftriaxone | 133 (80.6) | 13 (37.1) | 82 (78.8) | 13 (36.1) | 215 (79.9) | 26 (36.6) |
| Ceftazidime | 127 (76.9) | 11 (31.4) | 83 (79.8) | 12 (33.3) | 210 (78) | 23 (32.4) |
| Cefepime | 83 (50.3) | 17 (48.6) | 86 (82.6) | 12 (33.3) | 169 (62.8) | 29 (40.8) |
| Cefotaxime | 106 (64.2) | 10 (28.6) | 82 (78.8) | 11 (30.5) | 188 (69.9) | 21 (29.6) |
| Cefuroxime | 132 (80) | 19 (54.3) | 77 (74) | 17 (47.2) | 209 (77.7) | 36 (50.7) |
| Ciprofloxacin | 137 (83) | 27 (77.1) | 89 (85.5) | 19 (52.8) | 226 (84) | 46 (64.8) |
| Colistin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Cotrimoxazole | 120 (72.7) | 23 (65.7) | 82 (78.8) | 18 (50) | 202 (75) | 41 (57.7) |
| Ertapenem | 57 (34.5) | 5 (14.2) | 61 (58.6) | 15 (41.6) | 118 (43.9) | 20 (28.2) |
| Gentamicin | 59 (35.7) | 9 (25.7) | 55 (52.8) | 7 (19.4) | 114 (42.4) | 16 (22.5) |
| Imipenem | 88 (53.3) | 11 (31.4) | 70 (67.3) | 14 (38.9) | 158 (58.7) | 25 (35.2) |
| Levofloxacin | 128 (77.5) | 16 (45.7) | 69 (66.3) | 23 (63.8) | 197 (73.2) | 39 (54.9) |
| Meropenem | 136 (82.4) | 7 (20) | 93 (89.4) | 9 (25) | 229 (85.1) | 16 (22.5) |
| Norfloxacin | 146 (88.4) | 12 (34.3) | 83 (79.8) | 13 (36.1) | 229 (85.1) | 25 (35.2) |
| Nitrofurantoin | 26 (15.7) | 4 (11.4) | 81 (77.9) | 7 (19.4) | 107 (39.8) | 11 (15.5) |
| Piperacillin-tazobactum | 53 (32.1) | 7 (20) | 41 (39.4) | 5 (13.8) | 94 (34.9) | 12 (16.9) |
Molecular Analysis by PCR
The overall incidence of β-lactamase genes was found to be 72.6% (247 out of 340) which included 78% (156 out of 200) of E. coli and 65% (91 of 140) of K. pneumoniae, respectively. Molecular characterization showed that out of the 269 phenotypically positive ESBL isolates, 31.97% (86 out of 269) were positive for the blaCTX-M1 group (►Fig. 2, lane 3, 4, 5, 6, 8). It included 29% (48 out of 165) of E. coli and 36.5% (38 out of 104) of K. pneumoniae. Overall blaTEM (49.4%) was the commonest genotype followed by blaCTX-M1 (31.97%), blaOXA-1 (37.5%), and blaSHV (11.9%) either alone or in combination. In CTX-M1 cluster, 84.9% (73 out of 86) belonged to CTX-M15. In total, 2.6 and 5.2% of the isolates were positive for PER-2 and VEB, respectively. None of the isolates harbored GES gene (►Table 4). In total, 35% of the total ESBL-positive isolates harbored the three ESBL genes, while 61% carried two of the tested ESBL genes.
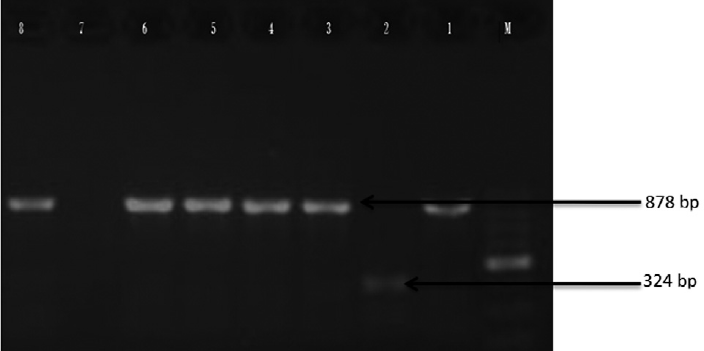
- Agarose gel electrophoresis of products obtained by multiplex PCR. Lane M: 100 bp DNA ladder; L1: Positive control for blaM-1 cluster (878 bp); L2: Positive control for blaM-2 cluster (324 bp); L3: E. coli isolate positive for blaM1 gene (874 bp); L4: K. pneumoniae isolate positive for blaM1 gene (878 bp). PCR, polymerase chain reaction.
| Escherichia coli (N = 165) (%) | Klebsiella pneumoniae N = 104) (%) | Total (N = 269) (%) | |
|---|---|---|---|
| CTX M-1 | 48 (29) | 38 (36.5) | 86 (31.97) |
| CTX M-2 | 3 (1.82) | 1 (0.96) | 4 (1.49) |
| CTX M-9 | 2 (1.21) | 0 (0) | 2 (0.74) |
| CTX- M-15 | 39 (23.6) | 34 (32.7) | 73 (27.1) |
| TEM | 78 (47.2) | 55 (52.9) | 133 49.4) |
| SHV | 18 (10.9) | 14 (13.5) | 32 (11.9) |
| OXA-1 | 54 (32.78) | 27 (25.9) | 81 (30.1) |
| VEB | 9 (5.4) | 5 (4.8) | 14 (5.2) |
| PER-2 | 6 (3.6) | 1 (0.96) | 7 (2.6) |
| GES | 0 (0) | 0 (0) | 0 (0) |
Abbreviations: ESBL, extended-spectrum β lactamase.
Discussion
Urinary tract infections are a highly prevalent and major issue among hospitalized patients, and multidrug-resistant E. coli and K. pneumoniae are the most important pathogens causing HAUTIs. ESBLs resistance in these strains poses a serious therapeutic challenge to clinicians as there are limited treatment options for the infections caused by ESBL-producing bacteria. The identification of the genes involved in ESBL-mediated resistance is usually done for surveillance, epidemiological studies, prevention of dissemination of resistance in hospital, and infection control.[10] In our study, the overall prevalence of ESBL production was 79.1%, which included 82.5% isolates of E. coli and 74.3% isolates of K. pneumoniae, which was higher when compared with other studies. ESBL production in various studies in India reported a prevalence of 60.8, 46.25, 66.77, 33, 63.6, and 18.8%, and 39.2, 25, 61.7, 42, 66.7, and 35.5% in E. coli and K. pneumoniae isolates, respectively.[2,10-15] The worldwide prevalence of ESBL production among the clinical isolates is found to be ranging from less than 1 to 88%.[5,9,13]
In our study, the highest rate of susceptibility was found to colistin followed by piperacillin and tazobactam, nitrofurantoin, amikacin, gentamicin, and ertapenem. Studies have found colistin to be effective for the treatment of UTI caused by MDR gram-negative bacteria but colistin should be used judiciously due to its potential to cause nephrotoxicity.[16] Thus, nitrofurantoin, piperacillin and tazobactam, and aminoglycosides may be used in empiric treatment, especially in critical care settings. Carbapenems are usually the drugs of choice for the treatment of the infections caused by ESBL-producing bacteria but we found high resistance in meropenem and imipenem which is a matter of concern. Both the isolates in study showed high coresistance to other non-β-lactam antibiotics which is alarming and such situation poses difficulty in the management of HA-UTI ESBL infections. Only colistin exhibited 100% activity in both isolates. This is extremely alarming and calls for the judicious use of colistin especially in India. Few similar studies have reported colistin as the most sensitive drug for both E. coli and K. pneumoniae which is concurrent with our findings but reported higher carbapenem susceptibility.[2,17] A retrospective cross-sectional study in South India reported high resistance among uropathogens to third-generation cephalosporins, trimethoprim and sulfamethoxazole, amoxicillin-clavulanic acid, and fluoroquinolones and better sensitivity to aminoglycosides, and piperacillin-tazobactam which is similar to our study.[12] Surveillance of antimicrobial resistance isolates from seven hospitals across India showed colistin and carbapenems as the most sensitive drugs followed by amikacin and piperacillin/tazobactam.[13] A study for monitoring antimicrobial resistance trends for ESBL production in the Asia-Pacific region reported higher resistance to ciprofloxacin, levofloxacin, and most β-lactams, with the exception of amikacin, piperacillin/tazobactam, imipenem, and ertapenem in E. coli and K. pneumoniae.[1] A similar study showed significantly lower resistance to cephalosporins (58%), cotrimoxazole (59%), piperacillin and tazobactam (23%), carbapenems (5.9%), and norfloxacin (48%), compared with our study.[10] The higher incidence of antibiotic resistance among bacteria in India is largely due to the misuse of antibiotics. We found the incidence of multidrug-resistant bacteria to be E. coli (89%) and K. pneumoniae (91.4%). Different studies in India have reported the rates of multidrug resistance among E. coli to be ranging from 31 to 93% and among K. pneumoniae to be 20 to 95%.[2,10,13]
Data on various ESBL-producing genes in E. coli and K. pneumoniae in nosocomial urinary isolates by molecular methods are limited from India. In our study, overall incidence of β-lactamase genes was found to be 72.6% which included E. coli (78%) and K. pneumoniae (65%). Overall, blaTEM (49.4%) was the commonest genotype followed by blaCTX-M1 (31.97%), blaOXA-1 (30.1%), and blaSHV (11.9%) either alone or in combination. In total, 84.9% of the blaCTX-M1-positive isolates were also blaCTXM-15 positive. In total, 5.4 and 3.6% of the E. coli isolates and 4.8 and 0.96% of the K. pneumoniae were positive for VEB and PER-2 genes, respectively. None of the E. coli and K. pneumoniae harbored GES genes. In total, 35% of the total ESBL-positive isolates harbored the three ESBL genes, while 61% carried two of the tested ESBL genes. Our findings document evidence of the high prevalence of multidrug-resistant TEM-type-ESBL followed by CTX-M-1 and blaOXA-1 among nosocomial isolates in this region. This is a serious issue, and there is a possibility of community-based dissemination of ESBL-producing isolates. The high rate of various ESBL-producing genes poses significant implications for patient treatment and highlights the importance of strengthening antimicrobial surveillance, antibiotic stewardship, and regular monitoring of the rate of ESBL production along with multidrug resistance among nosocomial isolates.
In a multicentric study from tertiary care hospitals in India, E.coli isolates harbored blaTEM (50%), blaOXA-1 (43%), blaSHV (16%), and blaCTX-M2 (1%), which is similar to our study but the study reported a lesser rate of blaCTX-M1(21%) compared with our study and K. pneumoniae isolates 38, 29, 27, 21, and 0.6% harbored blaTEM, blaOXA-1, blaCTXM-1, blaSHV, and blaCTX-M2, respectively.[2] In Sikkim and Darjeeling, CTX-M15 was most prevalent followed by TEM, SHV, and OXA-2 ESBL genes.[18] Few studies from Central India varied from our study and reported blaTEM gene most predominant followed by blaCTX-M and blaSHV.[17,19,20] In a study from Assam, CTX-M, TEM, and SHV were detected in 54.4, 33.9, and 15.4% isolates, respectively.[21] In a study conducted in North East districts in India, blaCTX-M was identified in E. coli (88.67%) and blaTEM in K. pneumoniae (77.58%) as the most predominant genotype.[9] Roy et al found a higher rate of blaCTX-M15 (100%) compared with our study (37.5%).[22] In a study in southern India, E. coli (63.7%) and K. pneumoniae (36.2%) were positive for CTX-M-1.[11] Various investigators from different countries have reported varying data on the ESBL. The results of our study are slightly comparable to data available from previous publications from Jordan,[15] Sudan,[23] Japan,[24] Saudi Arabia,[25] Pakistan,[26] Iran,[27] Iraq,[28] Brazil,[29] and China,[30] wherein the CTX-M prevalence was 32.4 to 91.7%, blaTEM 28 to 81%, blaSHV 4.4 to 35%, blaOXA-1 3 to 54.8%, and blaVEB 0 to 10.6%, thus, indicating that ESBL-encoding genes have disseminated across the world.
VEB, PER-2, and GES types of ESBL remain underreported in Indian hospitals as molecular characterization is not routinely performed to screen them in diagnostic laboratories. In present study, 2.6 and 5.2% of the isolates were positive for PER-2 and VEB, respectively, in ESBL-producing E. coli and K. pneumoniae, and none of the isolates was positive for GES genes. In a study conducted among health care infection isolates in Thailand, bla genes-encoding VEB-1 was found in 8.5% ESBL-producing E. coli and 10.2% ESBL-producing K. pneumoniae.[30] A study in China found blaGES in 10% ESBL isolates, but blaPER, blaVEB, and blaOXA group genes were not detected.[4] A similar study from Iran found blaOXA-1 gene in 17.24% of UPEC isolates but did not detect blaPER-1 and blaVEB genes.[31] Our findings are similar to study from north Iran which found VEB gene in 8% and no GES genes in ESBL isolates.[3] A similar study conducted in India showed disagreement with our study and found blaVEB in only 2.61% ESBL isolates.[32]
Conclusion
The findings of the present study contribute to better knowledge of the epidemiology of ESBL among HAUTIs caused by E. coli and K. pneumoniae in north India. High antibiotic drug resistance to antibiotics poses a major challenge for the management of ESBL infections and compels clinicians to the increased usage of carbapenems and colistin. There are the high prevalence of TEM, CTX-M-1, CTX-M-15, and SHV-type ESBL and the emergence of minor variants like OXA-1, VEB, and PER-2 type β-lactamase. Dissemination of ESBL warrants the need for more detailed surveillance to adopt appropriate control measures. There is a serious need for infection control checklists and bundles to be implemented in hospitals. Local antibiograms are essential to guide the selection of suitable antibiotics for empirical treatments. The findings of this study will prove valuable in the development of treatment strategies against HAUTIs in India.
Acknowledgments
We would like to acknowledge Late Prof. T. N. Dhole, Professor, Department of Microbiology and laboratory staff who helped to carry out this study.
Conflict of Interest
None declared.
References
- Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010-2013. Int J Antimicrob Agents. 2016;47(04):328-334.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of extended-spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. Indian J Med Res. 2019;149(02):208-215.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of multidrug resistant extended-spectrum beta-lactamase-producing Escherichia coli among uropathogens of pediatrics in North of Iran. BioMed Res Int. 2015;2015 309478
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization and epidemiology of extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008;52(08):2818-2824.
- [CrossRef] [PubMed] [Google Scholar]
- Simple and reliable multiplex PCR assay for detection of blaTEM, bla(SHV) and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223(02):147-151.
- [CrossRef] [PubMed] [Google Scholar]
- Twenty-Second Informational Supplement: Clinical and Laboratory Standards Institute, (CLSI) Wayne, PA: Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing 2013:M100-S21.
- [Google Scholar]
- Multidrug-resistant ESBL-producing Enterobacteriaceae and associated risk factors in community infants in Lebanon. J Infect Dev Ctries. 2016;10(09):947-955.
- [CrossRef] [PubMed] [Google Scholar]
- Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(03):490-495.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of blaTEM, blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Indian J Pathol Microbiol. 2014;57(02):249-254.
- [CrossRef] [PubMed] [Google Scholar]
- Changing antibiotic susceptibility pattern in uropathogenic Escherichia coli over a period of 5 years in a tertiary care center. Infect Drug Resist. 2019;12:1439-1443.
- [CrossRef] [PubMed] [Google Scholar]
- Study of CTX-M type of extended spectrum β-lactamase among nosocomial isolates of Escherichia coli and Klebsiella pneumoniae in South India. Indian J Microbiol. 2012;52(01):35-40.
- [CrossRef] [PubMed] [Google Scholar]
- Extended-spectrum beta-lactamase-producing community-acquired urinary tract infections in children: chart review of risk factors. J Glob Infect Dis. 2018;10(04):222-225.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial susceptibility profiles of gram-negative bacteria causing infections collected across India during 2014-2016: study for monitoring antimicrobial resistance trend report. Indian J Med Microbiol. 2018;36(01):32-36.
- [CrossRef] [PubMed] [Google Scholar]
- Extended spectrum beta-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. Indian J Med Res. 2009;129(06):695-700.
- [Google Scholar]
- Antimicrobial Susceptibility of Klebsiella pneumoniae and Escherichia coli with extended-spectrum β-lactamase associated genes in hospital Tengku Ampuan Afzan, Kuantan, Pahang. Malays J Med Sci. 2016;23(02):14-20.
- [Google Scholar]
- Effectiveness and safety of colistin in multidrug resistant urinary tract infections. J Appl Pharm Sci. 2017;7(09):148-152.
- [Google Scholar]
- Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum β-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adults. 3 Biotech. 2017;7(04):244.
- [CrossRef] [PubMed] [Google Scholar]
- The first report of phenotypic and molecular characterization of extended-spectrum beta-lactamase-producing uropathogens in Sikkim and Darjeeling hills of India. Microb Drug Resist. 2018;24(09):1284-1288.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of TEM and CTX-M genes from ciprofloxacin resistant Proteus mirabilis and Escherichia coli isolated on urinary tract infections (UTIs) Microb Pathog. 2018;121:123-130.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of blaCTX-M sequences of Indian origin and thirteen uropathogenic Escherichia coli isolates resistant to multiple antibiotics. BMC Res Notes. 2018;11(01):630.
- [CrossRef] [PubMed] [Google Scholar]
- New Delhi metallo-β-lactamase and extended spectrum β-lactamases co-producing isolates are high in community-acquired urinary infections in Assam as detected by a novel multiplex polymerase chain reaction assay. Indian J Med Microbiol. 2016;34(02):173-182.
- [CrossRef] [PubMed] [Google Scholar]
- Neonatal septicaemia caused by diverse clones of Klebsiella pneumoniae & Escherichia coli harbouring blaCTX-M-15. Indian J Med Res. 2013;137(04):791-799.
- [Google Scholar]
- Prevalence of TEM, SHV and CTX-M genes in Escherichia coli and Klebsiellaspp. urinary isolates from Sudan with confirmed ESBL phenotype. Life Sci J. 2013;10:191-195.
- [Google Scholar]
- Molecular characteristics of extended-spectrum β-lactamases in clinical isolates from Escherichia coli at a Japanese tertiary hospital. PLoS One. 2013;8(05):e64359.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of carbapenemases, ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J Infect Public Health. 2018;11(01):64-68.
- [CrossRef] [PubMed] [Google Scholar]
- Distribution of blaCTX - M, bla TEM, bla SHV and bla OXA genes in extended-spectrum-β-lactamase-producing clinical isolates: a three-year multi-center study from Lahore, Pakistan. Antimicrob Resist Infect Control. 2019;8(01):80.
- [CrossRef] [PubMed] [Google Scholar]
- Antibiotic resistance, virulence and genetic diversity of Klebsiella pneumoniae in community- and hospital-acquired urinary tract infections in Iran. Acta Microbiol Immunol Hung. 2019;66(03):349-366.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterr J Hematol Infect Dis. 2019;11(01):e2019041.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of OXA-1 β-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS One. 2014;9(03):e91800.
- [CrossRef] [PubMed] [Google Scholar]
- Phenotypic and genotypic characterization of multi-drug-resistant Escherichia coli isolates harboring blaCTX-M group extended-spectrum β-lactamases recovered from pediatric patients in Shenzhen, southern China. Infect Drug Resist. 2019;12:1325-1332.
- [CrossRef] [PubMed] [Google Scholar]
- Genotyping of ESBL producing uropathogenic and diarrheagenic Escherichia coli in southeast of Iran. Infect Disord Drug Targets. 2015;15(02):118-124.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence and characterization of beta-lactamase-producing Escherichia coli isolates from a tertiary care hospital in India. J Lab Physicians. 2019;11(02):123-127.
- [CrossRef] [PubMed] [Google Scholar]





