Translate this page into:
Risk factors for intestinal colonization with vancomycin resistant enterococci’ A prospective study in a level III pediatric intensive care unit
Address for correspondence: Dr. Sujatha Sistla, Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry - 605 006, India. E-mail: sujathasistla@gmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
PURPOSE:
Vancomycin-resistant enterococci (VRE) emerged as one of the major nosocomial pathogens across the globe. Gut colonization rate with VRE is higher in patients admitted to intensive care units (ICUs) due to the higher antibiotic pressure. VRE colonization increases the risk of developing infection up to 5–10 folds. The aim of this study was to determine the rates of VRE colonization among the patients admitted to pediatric ICU (PICU) and risk factors associated with it.
MATERIALS AND METHODS:
Rectal swabs were collected after 48 h of admission to PICU from 198 patients. The samples were inoculated onto bile esculin sodium azide agar with 6 mg/ml of vancomycin. Growth on this medium was identified by the standard biochemical test, and minimum inhibitory concentration of vancomycin and teicoplanin was detected by agar dilution method. Resistance genes for vancomycin were detected by polymerase chain reaction. Risk factors were assessed by logistic regression analysis.
RESULTS:
The rates of VRE colonization in patients admitted to PICU was 18.6%. The majority of the isolates were Enterococcus faecium (75.6%) followed by Enterococcus faecalis (24.4%). One patient acquired a VRE bloodstream infection (2.6%) among colonized patients, and none of the noncolonized patients acquired the infection. Consumption of vancomycin was found to be the only risk factor significantly associated with VRE colonization.
CONCLUSION:
Routine surveillance and isolation of patients found to be VRE colonized may not be possible in tertiary care hospitals; however, educating health-care workers, promoting handwashing with antiseptic soaps or solutions, and antibiotic Stewardship policy may help in the reduction of vancomycin resistance and VRE colonization.
Keywords
Enterococcus faecalis
Enterococcus faecium
vancomycin-resistant enterococci
vancomycin-resistant enterococci
vancomycin-resistant enterococci colonization in Intensive Care Unit
Introduction
After the first report of vancomycin-resistant enterococci (VRE) was published from the UK and France in 1986, it has emerged as one of the major nosocomial pathogens across the globe.[1] Long-term stay in the hospitals may result in VRE colonization in the gut thus creating reservoirs of infection. Transmission of VRE from patient to patient occurs through hands of health-care workers and through contaminated fomites in the environment.[2] VRE colonization increases the risk of developing infection up to 5–10 fold.[3]
Enterococcus faecalis and Enterococcus faecium contribute to 80%–90% of all enterococcal infections such as bloodstream infection, endocarditis, meningitis, urinary tract infection, and wound infection.[4] Multidrug resistance, increased rate of gut colonization, use of broad-spectrum antibiotics, and invasive devices have been implicated in the emergence of VRE.[5] VRE bacteremia usually results in increased mortality rate and treatment cost when compared to bacteremia caused by vancomycin-sensitive enterococci.[67]
VRE colonization is a major burden in intensive care units (ICU's) of large tertiary care hospitals. The higher antibiotic pressure in the ICU's contributes to the increased rate of VRE colonization. Once patients get colonized with VRE, it may persist from 7 weeks to 3 years, which may lead to a variety of infections in ICU patients.[8] The risk factors, namely, prolonged hospitalization, immunosuppression, low birth weight, younger age group, and prolonged antibiotic consumption of vancomycin, teicoplanin, amikacin, and ceftazidime were found to be associated with VRE colonization in pediatric and newborn ICUs and also in children's hospitals.[59101112131415]
There are limited reports of VRE colonization particularly from pediatric ICU (PICU) across the globe with rates ranging from 0% to 66%.[2591617181920212223] A study from our neighboring country Pakistan (Southeast Asia) reported a VRE colonization in PICU of 10.7%.[5] At the time of undertaking this study, to the best of our knowledge, no reports were available from India on VRE colonization in PICU's. The main objectives of this study were to determine the rate of VRE colonization in patients admitted to PICU and to assess the risk factors associated with it.
Materials and Methods
Study design and subject
A prospective study was conducted in level III PICU of a tertiary care teaching hospital from September 2013 to February 2015. Written informed consent was obtained from parents/guardians of the admitted patients, and the study was approved by the Institutional Ethics Committee.
Intervention/data collection
A structured pro forma was used to document the patient's demographic and clinical details, which included age, sex, medical history, clinical diagnosis, prior hospital/ICU admission, date of present admission to hospital and PICU, and history of antibiotic usage.
Outcome measures
-
Screening rates for VRE in patients admitted to PICU
-
Risk factors associated with VRE colonization.
Sample size
The sample size was calculated to be 196 using open Epi Version 3.01 (Armonk, NY, IBM Corp), taking the 17% positivity rate from a pilot study with 5% absolute precision, 5% alpha error, and 10% nongrowth or contaminated samples.
Inclusion criteria
The patients who were admitted for >48 h in the ICU with the consent from parents/guardians were included in the study.
Screening and diagnostic procedures
During the study period, 1290 patients were admitted in the PICU, out of which 1018 patients stayed >48 h. As calculated sample size was 196, convenient sampling was carried out to achieve the sample size. Rectal swabs were collected after 48 h of admission to PICU and inoculated onto bile esculin sodium azide agar containing 6mg/L vancomycin (Himedia laboratories, Mumbai, India).[5] Black/brown colonies were presumptively identified as Enterococcus and confirmed to species level based on Facklam and Collins standard biochemical tests.[24] Minimum inhibitory concentration (MIC) of vancomycin and teicoplanin was determined by agar dilution method for all the enterococcal isolates grown on BEA as per the Clinical and Laboratory Standard Institute guidelines.[25] MIC of ≥32 was considered as resistant to both vancomycin and teicoplanin. The antimicrobial susceptibility of the isolates to other antibiotics, namely, ampicillin, high-level gentamicin, tetracycline, and linezolid was also performed by Kirby-Bauer disc diffusion method.[26] Association of VRE colonization with demographic features of the patients and various risk factors for VRE colonization were assessed based on previous studies.
Molecular detection of vancomycin resistance genes
Genomic DNA was extracted from all the phenotypically vancomycin-resistant isolates using Qiagen mericon DNA extraction kit as per the manufacturer instructions. Polymerase chain reaction (PCR) was carried out to detect the vancomycin-resistance genes, namely, vanA, vanB, vanC-1, vanC-2, and vanC-3, using published primers.[27] PCR was done in a Mastercycler nexus (Eppendorf) with initial denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 1 min, annealing temperature at 54°C for 1 min, and extension at 72°C for 1 min, followed by the final extension at 72°C for 10 min. PCR products were run on 1.5% agarose gel with 100 bp ladder, stained with ethidium bromide and visualized under Gel Doc XR + System (Bio-Rad Laboratories).
Statistical analysis
Data were analyzed using SPSS 20.0 version software (OpenEpi: Open Source Epidemiologic Statistics for Public Health, Version. www.OpenEpi.com). The outcome of colonization with VRE was expressed as a binary categorical variable. All categorical variables were analyzed using Chi-square test while logistic regression was used for continuous independent variables and categorical outcome. The P < 0.05 was considered statistically significant. The difference in difference test was used to analyze the difference between the proportions of VRE-positive cases in PICU.
Results
Out of 1290 patients admitted in PICU during the study period, 198 were screened for VRE. The detailed flow chart is mentioned in Figure 1.
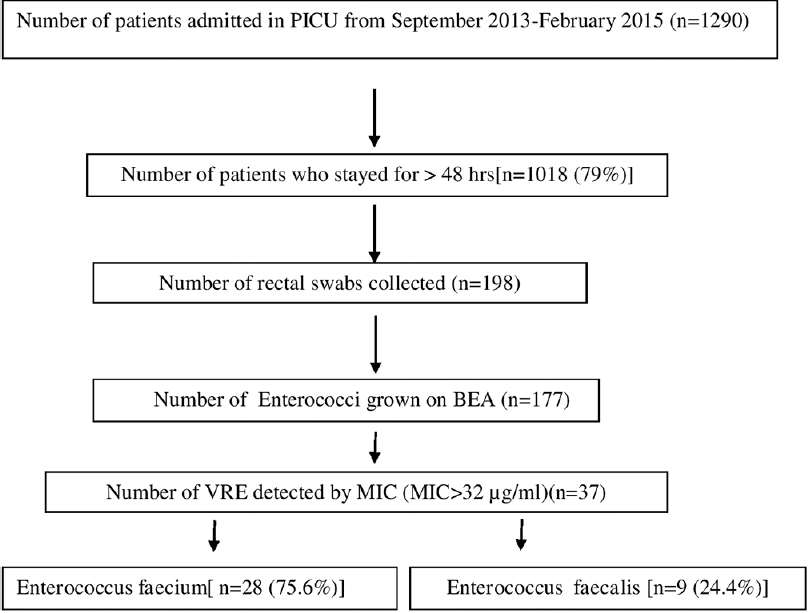
- Flow chart of study
The rectal swabs were collected from 198 patients admitted to PICU, out of which 37 (18.6%) yielded VRE. Of the VRE isolates, 75.6% were E. faecium and 24.4% were E. faecalis. The MIC values for both vancomycin and teicoplanin ranged from 64 to ≥256 for all the VRE isolates. Vancomycin resistance was always associated with teicoplanin resistance indicating that these VRE isolates were vanA-phenotype. PCR also confirmed vanA gene in all the 37 VRE isolates and none of them were positive for vanB and vanC genes. Very high rates of resistance to other antibiotics were noted with disc diffusion method (89%, 82%, and 86% to ampicillin, high-level gentamicin, and tetracycline, respectively). No resistance was observed to linezolid.
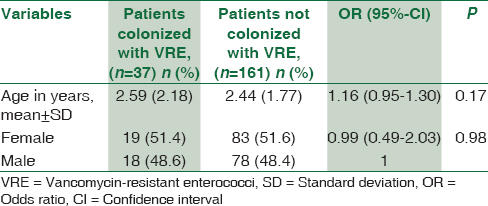
Among the various risk factors assessed, none of the demographic features of the patients admitted to PICU were associated with VRE Colonization [Table 1]. The average patient stay in the PICU before sample collection was 3.5 days (standard deviation ±3.84). No statistically significant association was found with VRE colonization rate and duration of patient stay, history of (H/O) previous hospital stay, and H/O surgery or urinary catheterization in PICU. Among the various antibiotics, vancomycin consumption was the only risk factor found to be significantly associated with VRE colonization in patients admitted to PICU (P = 0.006) [Table 2]. The average duration of vancomycin consumption was 2.2 days among VRE colonizers and 1.9 days among noncolonizers before sample collection. Of the 37 colonized patients, one patient acquired bloodstream infection after 7 days of VRE colonization. On the other hand, none of the 161 noncolonized patients acquired VRE infection.

Discussion
This study was conducted to detect the rates of VRE colonization among the patients admitted to PICU in a larger tertiary care hospital in southern India. ICUs are the major pool of infections with amplifying antimicrobial resistance. The heavy antibiotic burden in the ICUs provide a favorable environment for the development of resistance.[28] In this study, the rate of gut colonization with VRE was found to be 18.6% among patients admitted to PICU. In a study by Yameen et al., 2013, from Pakistan, 10.7% of VRE colonization rate was reported from patients admitted to PICU.[5] Nateghian et al., 2014,[23] from Iran, reported a high incidence of VRE colonization (66%). The rate of VRE colonization (18.6%) in our study correlates with other studies conducted in pediatric hospitals.[3202122] However, these studies also included patients admitted to pediatric wards in addition to ICUs. A few studies that include NICU patients reported higher rates of VRE colonization ranging from 39% to 66%[2923] whereas in a study reported by Bakir Saygan et al.,2010[18] and Singh et al., 2005,[19] from NICU, lower rates of VRE colonization of 0% and 3.5%, respectively, were encountered.
A meta-analysis study comprising adult patients admitted to hospitals in the USA reported an average VRE colonization rate of 12.5% with a range of 0%–42%.[29] However, data obtained from all the ICU's, that is, neonatal, pediatrics, and adults, VRE colonization rate varies from 0% to 66%.[212223272829] By comparing all these studies, it is clear that there is no specific association between age of the patients and colonization rates. These dissimilarities in the colonization rates may be dependent on geographical variation, hospital infection control policies, differences in health-care personal practices, and methodologies followed for the detection of colonization.
Previous studies have reported the association of various risk factors for VRE colonization such as prolonged hospitalization, immunosuppression, younger age group, prolonged antibiotic consumption such as vancomycin, teicoplanin, amikacin, and ceftazidime, administration of antimicrobial therapy for the late onset of neonatal sepsis.[159101112131415] In this study, the consumption of vancomycin was the only risk factor significantly associated with the VRE colonization. In contrast, in a study from our center in adult patients admitted to MICU, we observed prior ceftriaxone administration to be a significant risk factor.[30]
All the VRE isolates in this study had higher MIC value ranging from 64 ≥256 mg/L to both vancomycin and teicoplanin. This result depicts that all the strains are vanA-phenotype, that is, high-level resistance to both vancomycin and teicoplanin. Although the VRE isolates from this study showed very high rates of resistance to other antibiotics (89%, 82%, and 86% to ampicillin, high-level gentamicin, and tetracycline, respectively), no resistance was observed against linezolid. A few studies reported the emergence of resistance to linezolid among VRE isolates from colonized patients.[31] Antibiotics such as linezolid, tigecycline, and daptomycin are the treatment of choice for VRE infections, and most of the studies showed 100% sensitivity to these higher antibiotics.[32]
van A and van B are more commonly found among VRE isolates followed by vanC.[33] We also looked for the presence of van genes, that is, vanA, vanB, vanC-1, vanC-2, and vanC-3. In our study, only vanA gene was detected in all the VRE isolates, and no vanB and vanC genes were detected. vanA phenotypes are resistant to both vancomycin and teicoplanin due to the presence of vanA gene whereas vanB phenotype is only resistant vancomycin and sensitive to teicoplanin and carry vanB genotype. There is a variation in the global distribution of the van phenotype and genotypes. The majority of the VRE reported from the USA and Israel is the vanA phenotype while in Australia vanB phenotype is predominant.[34] In an earlier study from our hospital, we had reported both vanA and vanB phenotypes isolated from various clinical samples although in that study VRE isolates from both ICU and non-ICU patients were included.[4]
Out of 37 colonized patients in PICU, one patient (2.6%) acquired a VRE bloodstream infection after 7 days of detection of VRE colonization, while none of the 161 noncolonized patients was infected. VRE bloodstream infection rate among colonized patients ranging from 0% to 45% have been reported in many other studies.[3] The VRE isolated from rectal swab and blood of the same patient was of the same species, that is, E. faecium and exhibited similar antimicrobial sensitivity pattern. Based on these findings, we came to the purely tentative conclusion that the same strain went on to cause the infection. However, molecular characterization such as pulsed-field gel electrophoresis and multilocus sequence typing would have given a clearer picture about clonal relatedness.
The increase in VRE colonization and infection may lead to the risk of developing vancomycin resistance in Staphylococcus aureus strains. The colonization of patients with VRE and methicillin-resistant S. aureus (MRSA) increases the risk of acquiring vancomycin resistance by MRSA through plasmid-mediated gene transfer.[34] Hospital infection control practices advisory committee published the first guidelines in 1994 for the control of VRE infection in the hospitals.[35]
Limitation of the study is that rectal swab was collected at a single point of time. Follow-up rectal swabs of initially noncolonized patients would have provided information on VRE acquisition rate during the stay. We did not include pediatric control group in non-ICU areas which could have revealed any differences in the rates of VRE colonization in different hospital areas.
Conclusion
VRE colonization rate is moderately high in PICU of our hospital with prior vancomycin usage the only significant risk factor. Routine surveillance and isolation of patients found to be VRE-colonized may not be feasible in tertiary care hospitals; however, educating health-care workers, promoting handwashing with antiseptic soaps or solutions, and antibiotic Stewardship policy may help in the reduction of vancomycin resistance and VRE colonization.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors would like to thank JIPMER Intramural Funds, JIPMER, Puducherry.
References
- Characterization of a vancomycin-resistant Enterococcus faecium outbreak caused by 2 genetically different clones at a neonatal Intensive Care Unit. Ann Lab Med. 2012;32:82-6.
- [Google Scholar]
- Intervention to reduce transmission of resistant bacteria in intensive care. N Engl J Med. 2011;364:1407-18.
- [Google Scholar]
- Risk of vancomycin-resistant enterococci bloodstream infection among patients colonized with vancomycin-resistant enterococci. Braz J Infect Dis. 2015;19:58-61.
- [Google Scholar]
- Phenotypic & genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res. 2013;138:549-56.
- [Google Scholar]
- Nasal and perirectal colonization of vancomycin sensitive and resistant enterococci in patients of paediatrics ICU (PICU) of tertiary health care facilities. BMC Infect Dis. 2013;13:156.
- [Google Scholar]
- Driving forces of vancomycin-resistant E. faecium and E. faecalis blood-stream infections in children. Antimicrob Resist Infect Control. 2014;3:29.
- [Google Scholar]
- Outcomes associated with vancomycin-resistant enterococci: A meta-analysis. Infect Control Hosp Epidemiol. 2003;24:690-8.
- [Google Scholar]
- Vancomycin resistant enterococci healthcare associated infections. Ann Ig. 2013;25:485-92.
- [Google Scholar]
- Vancomycin-resistant Enterococcus colonization in neonatal Intensive Care Unit: Prevention and eradication experience. Mikrobiyol Bul. 2012;46:682-8.
- [Google Scholar]
- Vancomycin-resistant Enterococcus outbreak in a neonatal Intensive Care Unit: Epidemiology, molecular analysis and risk factors. Am J Infect Control. 2013;41:857-61.
- [Google Scholar]
- Prevalence of vancomycin-resistant enterococci among children with end-stage renal failure. Mid-European Pediatric Peritoneal Dialysis Study Group. Clin Infect Dis. 1999;29:912-6.
- [Google Scholar]
- Vancomycin-resistant Enterococcus colonization and infection in children: Six-year follow-up. Turk J Pediatr. 2014;56:618-25.
- [Google Scholar]
- Increased risk of vancomycin-resistant Enterococcus (VRE) infection among patients hospitalized for inflammatory bowel disease in the United States. Inflamm Bowel Dis. 2011;17:1338-42.
- [Google Scholar]
- Healthcare-associated vancomycin resistant Enterococcus faecium infections in the Mansoura University Hospitals Intensive Care Units, Egypt. Braz J Microbiol. 2015;46:777-83.
- [Google Scholar]
- Epidemiology of vancomycin-resistant enterococci in children with acute lymphoblastic leukemia at two referral centers in Tehran, Iran: A descriptive study. Int J Infect Dis. 2011;15:e332-5.
- [Google Scholar]
- Evaluation of vancomycin-resistant Enterococcus colonization at Gaziantep Children's Hospital, Turkey. Mikrobiyol Bul. 2011;45:646-54.
- [Google Scholar]
- Methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus carriage rates in a neonatal Intensive Care Unit. Mikrobiyol Bul. 2010;44:529-31.
- [Google Scholar]
- Control of vancomycin-resistant enterococci in the neonatal Intensive Care Unit. Infect Control Hosp Epidemiol. 2005;26:646-9.
- [Google Scholar]
- Epidemiology of enterococci in a neonatal Intensive Care Unit. Infect Control Hosp Epidemiol. 2008;29:374-6.
- [Google Scholar]
- The prevalence of vancomycin resistance genes in enterococci isolated from the stool of hospitalized patients in Mofid Children Hospital. Gene Ther Mol Biol. 2009;13:294-300.
- [Google Scholar]
- Control of a nosocomial outbreak of vancomycin resistant Enterococcus faecium in a paediatric oncology unit: Risk factors for colonisation. Eur J Pediatr. 1998;157:20-7.
- [Google Scholar]
- Prevalence of vancomycin resistant enterococci colonization and susceptibility to linezolid in pediatric Intensive Care Units of a referral pediatriccenter in Tehran, Iran. Arch Pediatr Infect Dis. 2014;2:e16970.
- [Google Scholar]
- Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731-4.
- [Google Scholar]
- Performance Standard for Antimicrobial Susceptibility Testing: 23rd Informational Supplement, M100-S23. Wayne: Clinical and Laboratory Standard Institute; 2013.
- [Google Scholar]
- Evaluating antimicrobial susceptibility test systems. In: Isenberg H, ed. Clinical Microbiology Procedures Handbook. Washington, DC: ASM Press; 1995. p. :1-14.
- [Google Scholar]
- Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:1434.
- [Google Scholar]
- Antibiotic stewardship programmes in Intensive Care Units: Why, how, and where are they leading us. World J Crit Care Med. 2015;4:13-28.
- [Google Scholar]
- Trends and significance of VRE colonization in the ICU: A meta-analysis of published studies. PLoS One. 2013;8:e75658.
- [Google Scholar]
- Screening for intestinal colonization with vancomycin resistant enterococci and associated risk factors among patients admitted to an adult Intensive Care Unit of a large teaching hospital. J Clin Diagn Res. 2016;10:DC06-9.
- [Google Scholar]
- Emergence of linezolid resistance in the vancomycin-resistant Enterococcus faecium multilocus sequence typing C1 epidemic lineage. J Clin Microbiol. 2006;44:1153-5.
- [Google Scholar]
- Antimicrobial susceptibility pattern of vancomycin resistant enterococci to newer antimicrobial agents. Indian J Med Res. 2015;141:483-6.
- [Google Scholar]
- Glycopeptide resistance in gram-positive cocci: A review. Interdiscip Perspect Infect Dis. 2012;2012:781679.
- [Google Scholar]
- VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:4580-7.
- [Google Scholar]
- 1995. Morbidity and Mortality weekly report. Recommendations for Preventing the Spread of Vancomycin Resistance. No. RR-12. 44 Available from: http://www.cdc.gov/mmwr/pdf/rr/rr4412.pdf





