Translate this page into:
The Prevalence and Antimicrobial Susceptibility Pattern of Gram-Positive Pathogens: Three-Year Study at a Tertiary Care Hospital in Mumbai, India
Address for correspondence: Sweta Shah, MD, Department of Laboratory Medicine, Kokilaben Dhirubhai Ambani Hospital and Research Institute, Four Bungalows, Andheri West, Mumbai, Maharashtra 400053, India (e-mail: shahsweta.microbiology@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction
The growing resistance pattern of the gram-positive pathogens along with a steady increase in minimum inhibitory concentration of the currently available antibiotics have led to an increase in morbidity and mortality rates in India. This study aims to access the shifting antibiotic susceptibility paradigm of the gram-positive pathogens in various infections at a tertiary care center.
Methods
This is a 3-year retrospective observational study which was performed from January 2016 to December 2018 at a tertiary care hospital in Mumbai. All clinically significant gram-positive cocci isolated from a variety of clinical specimens were studied for their prevalence and antimicrobial susceptibility.
Results
Out of 4,428 gram-positive isolates, Staphylococcus aureus (35.3%) was the commonly encountered pathogen, followed by Enterococcus spp. (32.1%) and coagulase-negative Staphylococcus (CoNS) (25.7%). S. aureus was majorly isolated from skin and soft tissue infections (60.3%), followed by patients with respiratory tract infections (18.2%) and blood stream infections (13%). Among S. aureus, particularly methicillin-resistant S. aureus (MRSA), prevalence increased from 29.5% in 2016 to 35.1% in 2018, with an overall prevalence of 33.6%. All S. aureus isolates were 100% sensitive toward vancomycin, linezolid, tigecycline, and teicoplanin. However, the CoNS isolates showed a higher resistance rate with reduced susceptibility toward linezolid and teicoplanin. High prevalence of resistance was observed across gram-positive isolates with commonly used antibiotics such as ciprofloxacin, levofloxacin, and erythromycin. While the prevalence of linezolid-resistant enterococcus (LRE) was 3.6%, vancomycin (VRE) and teicoplanin resistance among the enterococcus species was as high as 7.7% and 7.5%, respectively.
Conclusion
Rising methicillin resistance among the Staphylococcal species (MRSA and MR-CoNS) along with reduced susceptibility toward currently available anti-MRSA agents is a matter of serious concern as it limits the therapeutic options for treating multidrug resistant (MDR) gram-positive infections.
Keywords
gram-positive pathogens
Staphylococcus aureus
vancomycin-resistant enterococcus
coagulase-negative staphylococci
MRSA
Introduction
Antimicrobial resistance (AMR) is a serious global threat that alone would be responsible for 10 million deaths by 2050 and over USD 100 trillion economic losses if effective interventions against resistance are not made.[1] India has been termed as the “AMR capital” of the world.[2] Poor sanitary conditions, absence of effective infection-control policies, antibiotic overuse in humans and animals, and lack of awareness among the masses about the infectious diseases[3] have led to a gradual increase in AMR over time. With 10.7 units being consumed per person, India is the highest antibiotic consumer in the world.[4] Among the low- and middle-income countries, India alone accounted to an increase of 103% in antibiotic consumption from 2000 to 2015.[5]
Gram-positive pathogens play a critical role in health care–associated and community-acquired infections due to its rapid ability to acquire resistance against the currently used antibiotics. Prolonged usage of the available effective antibiotics against these pathogens coupled with their decreasing susceptibility rates have led to the emergence and spread of multidrug resistant (MDR) Staphylococcus and Enterococcus spp., particularly methicillin-resistant S. aureus (MRSA), vancomycin-resistant S. aureus (VRSA), and vancomycin-resistant Enterococcus (VRE). MRSA is considered a life-threatening nosocomial pathogen as it is associated with worse clinical outcomes[6] and increased complications[7] than patients with methicillin-sensitive strains (MSSA), leading to a higher mortality rate.[8] According to a recent Indian Council of Medical Research–Antimicrobial Resistance Surveillance Network (ICMR–AMRSN) report, the total MRSA prevalence in India escalated from 32.9% in 2017 to 38.6% in 2018, with North India reporting the highest MRSA prevalence of 52.8%, followed by West India (48.1%).[9]
Currently, the resistance rate of MRSA has significantly increased to non–β lactam antibiotics like ciprofloxacin, levofloxacin, and clindamycin as compared with MSSA isolates. Similarly, increasing minimum inhibitory concentration (MIC) to vancomycin, which has been a traditional “standard” therapy for the treatment of MDR gram-positive pathogens, has led to an inception of vancomycin intermediate S. aureus (VISA), VRSA, and VRE isolates. Therefore, the growing resistance pattern, mortality rate, increased burden on health care and community settings along with a shrinking antibiotic pipeline, has lead WHO list MRSA and VRSA as a “high”-priority pathogen against which novel and effective antibiotics are needed.[10]
The shifting antibiotic paradigms and the need to select targeted antibiotic regimen against resistant pathogens prompted us to investigate the prevalence and the susceptibility pattern of the gram-positive pathogens in various infections at our tertiary care center.
Methods
This is a 3-year retrospective analysis of the specimens received for aerobic culture and sensitivity from January 2016 to December 2018 at a tertiary care hospital in Mumbai, India. The samples were processed using standard microbiological techniques. All the samples were plated on 5% sheep blood agar, Mac Conkey's agar. Chocolate agar was used for respiratory samples and body fluids like cerebrospinal fluid (CSF). Sterile body fluids and tissue were also inoculated in automated blood culture bottle for enrichment. After doing gram staining and manual tests like catalase and coagulase, identification and sensitivity were done using Vitek2 (BioMérieux, France). The antibiotic susceptibility results were reported as per the Clinical and Laboratory Standards Institute (CLSI) guidelines.[11,12,13] All pathogens were reported with due clinical correlation. The commensals were reported only when they were grown in another specimen collected at a different time or from a different site.
Results
During the period of 3 years, a total of 4,428 gram-positive isolates were analyzed, out of which Staphylococcus aureus (35.3%) was the most dominant pathogen, followed by Enterococcus spp. (32.1%), coagulase-negative Staphylococcus (CoNS) (25.7%), and Streptococcus spp. (6.7%).
Gram-positive Isolates
A. aureus was majorly isolated from skin and soft tissue infections (60.3%) while it also represented 18.2% and 13% in respiratory tract infections (RTI) and blood stream infections (BSI), respectively; 4.6% of S. aureus isolates were from urinary tract infections (UTI).
CoNS were the major pathogens isolated from BSI (42.6%). While 40.5% of the CoNS pathogen were responsible for skin and soft tissue infections, only 4% and 3% were observed in RTI and UTI, respectively. Staphylococcus epidermidis (53.4%) was the most common species of CoNS, followed by Staphylococcus haemolyticus (25.8%), Staphylococcus hominis (14%), and Staphylococcus capitis (6.7%). Among CoNS, the most dominant pathogen in skin and soft tissue infections (SSTIs) and BSI was S. epidermidis (56.2% and 50.2%). Apart from S. epidermidis, both S. haemolyticus (19.7%) and S. hominis (20%) were observed to contribute equally in the development of BSI. In UTI, S. haemolyticus (72.7%) was the most common species of CoNS.
The Enterococcus spp. were the dominant causative pathogen behind UTI (51.5%), followed by SSTIs (26%), while only 9% and 6.7% were responsible for BSI and RTI, respectively. Both Enterococcus faecalis (49%) and Enterococcus faecium (51%) were equally prevalent among the Enterococcus spp. ►Table 1 depicts the total prevalence of S. aureus, CoNS, and Enterococcus spp.
| Infection | SSTIs | Respiratory tract infection | Blood stream infection | Urinary tract infection |
|---|---|---|---|---|
| Pathogen | ||||
| Staphylococcus aureus (n = 1,566) | 944, 60.3% | 285, 18.2% | 204, 13% | Nil |
| Coagulase-negative Staphylococcus (CoNS) (n = 1,141) | 486, 42.6% | Nil | 462, 40.5% | Nil |
| Enterococcus spp. (n = 1,422) | 370, 26% | Nil | 128, 9% | 733, 51.5% |
Among Streptococcus species, Streptococcus agalactiae (37.1%) was the most common isolate, followed by Streptococcus pyogenes (35.4%) and Streptococcus pneumoniae (14%).
Antimicrobial Susceptibility Pattern Among the Gram-Positive Isolates
An antibiotic sensitivity pattern for ciprofloxacin, erythromycin, clindamycin, tetracycline, levofloxacin, gentamycin, linezolid, vancomycin, daptomycin, tigecycline, teicoplanin, benzylpenicillin, and trimethoprim (TMX)/sulfamethoxazole (SMZ) were determined for Staphylococcal spp., Enterococcus spp., and Streptococcus spp.
The overall prevalence of MRSA over the period of 3 years was 33.6% (►Table 2). Moreover, the MRSA prevalence steadily increased from 29.5% in 2016 to 35.1% in 2018.
| Total S. aureus isolates | 2016 (n = 386) | 2017 (n = 783) | 2018 (n = 393) | Total prevalence | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| MRSA | 114 | 29.5 | 274 | 35 | 138 | 35.1 | 33.6% |
| MSSA | 272 | 70.4 | 509 | 65 | 255 | 64.9 | 66.3% |
►Fig. 1 shows the total sensitivity pattern of S. aureus isolates against various tested antimicrobials. The S. aureus isolates showed lower sensitivity toward ciprofloxacin (11.1%), levofloxacin (11.8%), erythromycin (46.1%), benzylpenicillin (0.9%), TMX/SMZ (61.3%), clindamycin (70.9%), and gentamycin (82.4%). All the S. aureus isolates were found to be highly sensitive toward vancomycin, linezolid, tigecycline, and teicoplanin (100%). Less than 1% of the strains were found to be sensitive to benzyl penicillin.
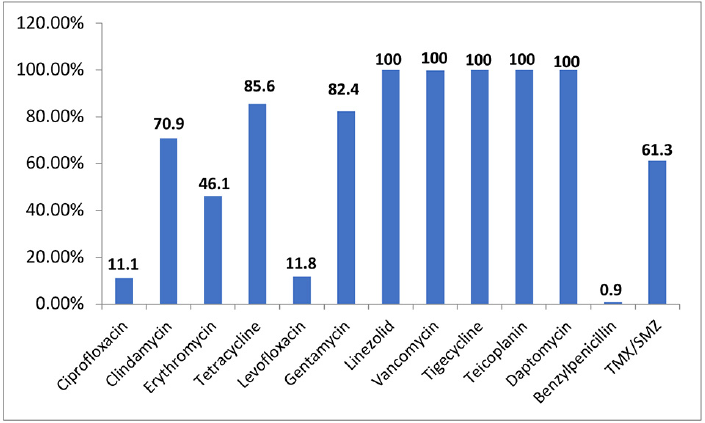
- Antibiotic sensitivity pattern among S. aureus isolates.
The prevalence of methicillin resistance among CoNS was 78.7%. ►Fig. 2 shows the sensitivity pattern in the CoNS isolates against various antibiotics tested. In this study, vancomycin remained highly sensitive toward the CoNS species as no vancomycin-resistant (VR-CoNS) isolate was documented. There was only one isolate of CoNS which showed intermediate susceptibility (VI-CoNS). The total teicoplanin sensitivity was 98.2% wherein 8 isolates were resistant and 12 isolates were teicoplanin intermediate. Four isolates of linezolid-resistant CoNS (LR-CoNS) were also encountered. The total resistance prevalence was observed to be higher in S. haemolyticus than S. epidermidis and S. hominis. In S. haemolyticus, resistance to cefoxitin was 90.5%, ciprofloxacin 82.2%, levofloxacin 70.2%, erythromycin 91.9%, and clindamycin 67.3%, while in S. epidermidis, the resistance to cefoxitin was 82.2%, ciprofloxacin was 53.2%, levofloxacin 39.6%, erythromycin 77%, and clindamycin in 55.6% of the isolates.
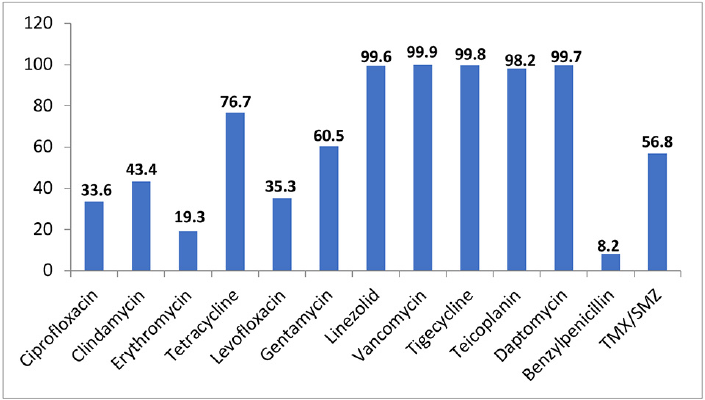
- Total sensitivity pattern of CoNS isolates against various tested antibiotics.
Although the prevalence of E. faecium and E. faecalis is similar, a decrease in susceptibility among the antimicrobials is more dominant in E. faecium than in E. faecalis (►Fig. 3). E. faecium demonstrated increased resistance in ciprofloxacin (92.9%), levofloxacin (92.4%), erythromycin (87.8%), and gentamycin (76.1%), as compared with E. faecalis which showed a resistance rate of 65.1% in ciprofloxacin, 58.7% in levofloxacin, 66.6% in erythromycin, and 49.4% in gentamycin. Further, vancomycin (VRE) and teicoplanin resistance was as high as 14.6% and 14.4%, respectively, in E. faecium. Linezolid-resistant enterococcus (LRE) was also found to be 3.6%. Tigecycline was highly sensitive to Enterococcus spp. except one isolate in E. faecium group that was found to be resistant. ►Fig. 3 elucidates the resistance pattern against the tested antimicrobials in both E. faecium and E. faecalis.

- Total antibiotic resistance pattern of Enterococcus species.
The streptococcus species were highly sensitive (100%) to vancomycin, teicoplanin, tigecycline, and linezolid. While Streptococcus species were susceptible to benzylpenicillin (90.5%) and ciprofloxacin (98%), it exhibited reduced sensitivity toward erythromycin (69.1%), tetracycline (41.7%), and moxifloxacin (75.8%).
Discussion
India has the highest burden of MDR pathogens, with Centre for Disease Dynamics, Economics and Policy (CDDEP) predicting two million deaths in India due to AMR by 2050.[14] WHO listed resistant gram-positive pathogens such as MRSA and VRSA as “high”-priority pathogens due to their ability to swiftly acquire resistance which eventually imposes an increased mortality burden.[10] Among the S. aureus, the overall MRSA prevalence in our study was 33.6% which was lower than studies from various centers across India where the prevalence rates vary from 38.6% to 52%.[15-17] Moreover, we observed that the MRSA prevalence rose from 29.5% in 2016 to 35% in 2018. This pattern was very similar to the recent reports which highlighted that over a decade, the MRSA prevalence has gradually escalated from 29% in 2009[18] to 39% in 2018.[15] This is a challenging scenario for clinicians as a recent study from south India reported that patients infected with MRSA have worse clinical outcomes in terms of longer hospital and intensive care unit (ICU) stay, greater number of complications, and increased mortality.[7]
Previous surveillance reports have shown S. aureus to be the major causative pathogen in SSTIs. Our study also reflected the same results with 60.3% of S. aureus being isolated from SSTIs caused by gram-positive organisms. Similar to previous studies, S. aureus isolates exhibited multidrug resistance as evident by their decreased sensitivity toward levofloxacin, ciprofloxacin, and erythromycin. An increase in the resistance rate against erythromycin is a huge concern for the clinicians as it mechanistically confers cross-resistance to clindamycin which is a preferred oral antibiotic for the treatment of SSTIs.
CoNS are a major nosocomial pathogen wherein S. epidermidis and S. haemolyticus are the most common species involved. CoNS have been reported to exhibit higher levels of MDR as compared with S. aureus, which makes their treatment challenging. The same thing is highlighted in our study where the methicillin resistance among CoNS was 78.7%, which is almost double than the methicillin resistance in S. aureus. Out of all the CoNS spp., S. haemolyticus has been shown to display the highest level of resistant antimicrobial pattern.[19,20] The ICMR-AMRSN study (2016–2018) has also shown the S. haemolyticus nonsusceptibility rate of 93% to penicillin and 81% to ciprofloxacin.[21] This pattern was reflected in our study also, where the prevalence of resistance encountered in S. haemolyticus was much higher than S. epidermidis.
Vancomycin is the choice of drug to treat MRSA infections. A surge in studies reporting vancomycin minimum inhibitory concentration (MIC) creep and reduced vancomycin susceptibility suggests its therapeutic limitation in severe infections. A recent study by Amberpet et al reported the rate of hVISA to be as high as 12.4%[22] while the prevalence of VRSA varies from 2 to 7.1% in India.[23,24] However, it was heartening to note that in our study, the Staphylococcal species were highly sensitive toward vancomycin with no resistant isolate being documented in S. aureus and CoNS isolate in a span of 3 years.
While there was no teicoplanin resistance among the S. aureus, CoNS demonstrated an evident rise in teicoplanin resistance (8 resistant and 12 intermediate isolates), especially in S. epidermidis isolates. These results were in line with previous studies[25-27] where teicoplanin-resistant CoNS species were reported. Though 98% of the CoNS isolates in our study are susceptible to glycopeptides, however, the rising methicillin and glycopeptide resistance among the CoNS species is a grave concern as it limits the currently available therapeutic options to treat MDR CoNS infections.
In India, the VRE strain first emerged in 1999.[28] Our study showed overall VRE prevalence of 7.7%, which was similar to previous reports from India where the prevalence varied from 2 to 11.9%.[21,29-31]E. faecium is associated with a higher antimicrobial resistant pattern and attributable mortality[21,32] as compared with E. faecalis. A recent study by Purohit et al reported that among the VRE prevalence (22.8%), the prevalence of VR E. faecium was as high as 33.9% while that of E. faecalis was 2.4%.[33] These isolates were also found to be resistant to teicoplanin. The ICMR–AMRSN report also emphasizes on the rising concern of the overall glycopeptide resistance which has escalated from < 15% to as high as 30%.[21] Although the rate of glycopeptide (vancomycin and teicoplanin) resistance was less in our study than the ICMR report, E. faecium displayed higher levels of glycopeptide resistance (14.5%) than E. facecalis (< 1%) and formed 98.1% of the total glycopeptide resistance in the Enterococcus spp.
Apart from vancomycin, linezolid is the most preferred drug which is approved for the treatment of resistant gram-positive infections. Linezolid was found to be highly sensitive against S. aureus and CoNS spp., wherein only four LR-CoNS were encountered. These observations reflect the Indian scenario with sporadic case studies which collectively report less than 1% of linezolid-resistant S. aureus isolates. The first report of linezolid resistant CoNS in India was reported in 2012,[34] after which there were many reports that document linezolid resistance in the CoNS species.[35-37] Recently, Mittal et al observed that the linezolid–teicoplanin resistant S. haemolyticus strain that was identified was not due to prior exposure to these antibiotics but due to the pathogen's ability to horizontally transmit resistant genes across species, thereby resulting in highly virulent nosocomial pathogen.[36]
Further, in India, the LRE isolates were first reported in 2014.[38] Our study showed high prevalence of LRE which was similar to a recent study from Mumbai which reported a high LRE prevalence of 4.1%.[31] Linezolid is the preferred drug in the treatment of infections caused by MRSA and VRE. Therefore, overuse of linezolid in patients where vancomycin treatment has failed might have led to an antibiotic selection pressure and subsequent co-occurrence of resistant strains.
In conclusion, this study demonstrates the steady increase in the emergence of highly resistant S. aureus, CoNS, and Enterococcus species. Although, in this study, a high efficacy against glycopeptides and linezolid was observed in the gram-positive isolates, however, the gradual increase in the resistance against these last line antibiotics for MDR gram-positive pathogens emphasizes the need for novel research antibiotics to combat these difficult-to-treat MDR pathogens. Therefore, this study provides an in-depth snapshot of the shifting susceptibility patterns of gram-positive pathogens, thereby facilitating the clinicians in choosing the appropriate antibiotic regimen which will help in reducing the selection pressure and subsequently controlling the rising AMR.
Authors’ Contribution
R.R. S.S: Data analysis, drafting of the manuscript, and critical revision. P.S., L.S.: Data generation and data entry. S.S., T.P.: Study concept and design, study supervision, data interpretation, critical revision, and final approval of the manuscript.
Conflicts of Interest
Authors Sweta Shah, Pooja Thakkar, Sushima Poojary, and Shweta Ladi have nothing to declare. Author Ritika Rampal is an employee of Wockhardt Limited.
References
- tackling a crisis for the health and wealth of nations. London: Welcome Trust; 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf. Accessed on May 20, 2020
- [Google Scholar]
- Antimicrobial resistance: the next big pandemic. Int J Community Med Public Health. 2017;4:2632-2636.
- [CrossRef] [Google Scholar]
- Antimicrobial resistance in the environment: the Indian scenario. Indian J Med Res. 2019;149(02):119-128.
- [CrossRef] [PubMed] [Google Scholar]
- Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14(08):742-750.
- [CrossRef] [PubMed] [Google Scholar]
- Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A. 2018;115(15):E3463-E3470.
- [CrossRef] [PubMed] [Google Scholar]
- Is methicillin-resistant Staphylococcus aureus more virulent than methicillin-susceptible S. aureus? A comparative cohort study of British patients with nosocomial infection and bacteremia. Clin Infect Dis. 2003;37(11):1453-1460.
- [CrossRef] [PubMed] [Google Scholar]
- Is methicillin-resistant Staphylococcus aureus infection associated with higher mortality and morbidity in hospitalized patients? A cohort study of 551 patients from South Western India. Risk Manag Healthc Policy. 2018;11:243-250.
- [CrossRef] [PubMed] [Google Scholar]
- National Treatment Guidelines for Antimicrobial Use in Infectious Diseases. New Delhi, India: Ministry of Health and Family Welfare, Government of India; 2016.
- [Google Scholar]
- 2018 https://www.icmr.nic.in/sites/default/files/reports/AMRSN_Annual_Report_2018_0.pdf. Accessed May 18, 2020
- WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(03):318-327.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI supplement M100S (26th). Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI supplement M100 (27th). Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. 28th Informational Supplement. In: CLSI Document M100. Wayne, PA: CLSI; 2018.
- [Google Scholar]
- State of the World's Antibiotics. Washington, DC: Center for Disease Dynamics, Economics and Policy; 2015. https://www.cddep.org/wp-content/uploads/2017/06/swa_edits_9.16.pdf. Accessed May 20, 2020
- [Google Scholar]
- http://iamrsn.icmr.org.in/index.php/resources/amr-icmr-data. Published 2018. Accessed May 28, 2020
- Methicillin resistance, vancomycin intermediate and vancomycin resistance Staphylococcus aureusprevalence in a tertiary care hospital of Punjab, India. National J Lab Med. 2019;8(03):MO01-MO03. DOI: 10.7860/NJLM/2019/41137:2352
- [Google Scholar]
- Recent pattern of antibiotic resistance in Staphylococcus aureus clinical isolates in Eastern India and the emergence of reduced susceptibility to vancomycin. J Lab Physicians. 2019;11(04):340-345.
- [CrossRef] [PubMed] [Google Scholar]
- Dos Santos KR, Giambiagi-Demarval M. Staphylococcus haemolyticus as an important hospital pathogen and carrier of methicillin resistance genes. J Clin Microbiol. 2012;50(01):166-168.
- [CrossRef] [PubMed] [Google Scholar]
- Whole-genome sequencing reveals clonal expansion of multiresistant Staphylococcus haemolyticus in European hospitals. J Antimicrob Chemother. 2014;69(11):2920-2927.
- [CrossRef] [PubMed] [Google Scholar]
- Establishing antimicrobial resistance surveillance and research network in India: journey so far. Indian J Med Res. 2019;149(02):164-179.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of heterogeneous vancomycin-intermediate Staphylococcus aureus: a preliminary report from south India. Indian J Med Res. 2019;150(02):194-198.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug-resistant Staphylococcus aureus, India, 2013-2015. Emerg Infect Dis. 2016;22(09):1666-1667.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J Med Res. 2011;134(05):704-708.
- [CrossRef] [PubMed] [Google Scholar]
- Teicoplanin non-susceptible coagulase-negative staphylococci in a large Australian healthcare network: implications for treatment with vancomycin. Sri Lankan Journal of Infectious Diseases. 2017;7(01):10-17.
- [CrossRef] [Google Scholar]
- Determination of vancomycin, teicoplanin and linezolid resistance among Staphylococcal isolates from a tertiary care hospital. J Acad Clin Microbiol. 2015;17:3-6.
- [CrossRef] [Google Scholar]
- Linezolid-resistant Staphylococcus spp. at a tertiary care hospital of Andhra Pradesh. Indian J Med Microbiol. 2011;29(03):314-315.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin-resistant Enterococcus bacteraemia in a lymphoma patient. Indian J Med Microbiol. 1999;17:194-195.
- [Google Scholar]
- Emergence of VRE and their antimicrobial sensitivity pattern in a tertiary care teaching hospital. J Med BiolSci. 2009;8:26-32.
- [Google Scholar]
- Phenotypic and genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res. 2013;138(04):549-556.
- [Google Scholar]
- Microbiological profile and antibiotic susceptibility pattern of gram-positive isolates at a tertiary care hospital. J Lab Physicians. 2019;11(02):144-148.
- [CrossRef] [PubMed] [Google Scholar]
- Vancomycin-resistant Enterococcal bacteremia: comparison of clinical features and outcome between Enterococcus faecium and Enterococcus faecalis. J Microbiol Immunol Infect. 2008;41(02):124-129.
- [Google Scholar]
- Characterization of vancomycin resistant Enterococci in hospitalized patients and role of gut colonization. J Clin Diagn Res. 2017;11(09):DC01-DC05.
- [CrossRef] [PubMed] [Google Scholar]
- Linezolid resistant Staphylococcus haemolyticus: first case report from India. Asian Pac J Trop Med. 2012;5(10):837-838.
- [CrossRef] [PubMed] [Google Scholar]
- Linezolid-resistant Staphylococcus haemolyticus: emergence of G2447U & C2534U mutations at the domain V of 23S ribosomal RNA gene in a tertiary care hospital in India. Indian J Med Res. 2019;149(06):795-798.
- [CrossRef] [PubMed] [Google Scholar]
- Linezolid resistant coagulase negative Staphylococci (LRCoNS) with novel mutations causing blood stream infections (BSI) in India. BMC Infect Dis. 2019;19(01):717.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanisms of linezolid resistance among coagulase-negative Staphylococci determined by whole-genome sequencing. MBio. 2014;5(03):e00894-e14.
- [CrossRef] [PubMed] [Google Scholar]
- The first linezolid-resistant Enterococcus faecium in India: high level resistance in a patient with no previous antibiotic exposure. Avicenna J Med. 2014;4(01):13-16.
- [CrossRef] [PubMed] [Google Scholar]





