Translate this page into:
Utility of hub and spoke model for estimating the burden of Group B Streptococcus infection and its antibiotic resistance profiling among pregnant tribal women
*Corresponding author: Shashank Purwar, Department of Microbiology, All India Institute of Medical Sciences, Bhopal, Madhya Pradesh, India. shashank.microbiology@aiimsbhopal.edu.in
-
Received: ,
Accepted: ,
How to cite this article: Gupta P, Roy L, Halder A, Mishra A, Yadav J, Pushpalatha K, et al. Utility of hub and spoke model for estimating the burden of Group B Streptococcus infection and its antibiotic resistance profiling among pregnant tribal women. J Lab Physicians. 2025;17:65-71. doi: 10.25259/JLP_178_2024
Abstract
Objectives
Group B Streptococcus (GBS) is the leading cause of maternal sepsis and neonatal meningitis. The objective is to determine the feasibility of using the hub-spoke model for the collection and transportation of recto-vaginal swab samples was evaluated in addition to the GBS burden among tribal females, convenient diagnostic method and antibiotic susceptibility profiling.
Materials and Methods
Recto-vaginal samples were collected from the 35–37 weeks pregnant women using pre-sterile HiCulture transport swab and were transferred into Lim Broth. Direct plating was carried out by inoculating swabs onto chromogenic agar. Positive colonies were subjected to antibiotic susceptibility testing using the Kirby–Bauer disc diffusion method. Polymerase chain reaction (PCR) was also performed.
Statistical analysis
Statistical tests were performed using Microsoft excel. P < 0.05 was considered statistically significant.
Results
The socio-demographic and clinical profiling of the 808 pregnant females suggested vaginal discharge (89.23%) to be the most common symptom at the time of presentation. GBS was detected in 116 (14.35%) and 52 (6.43%) by PCR and culture-based methods, respectively. The sensitivity and specificity of the PCR assay were estimated to be 100% and 91.53%, respectively. Obtained GBS isolates were found to be 100% sensitive to quinupristin – dalfopristin, linezolid, cefepime, and vancomycin, whereas 10%, 23%, 27%, and 23% of the isolates were found to be resistant for penicillin, clindamycin, erythromycin, and levofloxacin, respectively.
Conclusions
Hub-spoke model for GBS screening must be emphasized in rural settings to identify pregnant women requiring prophylactic antibiotic therapy. PCR could be used as an efficient tool for identifying GBS.
Keywords
Antibiotic susceptibility testing
Culture
Group B Streptococcus
Hub-spoke model
Neonatal mortality
Polymerase chain reaction
Pregnant females
Tribal area
INTRODUCTION
Group B Streptococcal infection or Group B streptococcal disease, caused by Streptococcus agalactiae, is Gram-positive cocci colonizing the lower gastrointestinal tract and has been known to be the leading cause of maternal sepsis and neonatal meningitis for decades.[1] Among neonates, usually two types of clinical presentations are reported by Group B Streptococcus (GBS) infection, that is, early-onset and late-onset disease. Early-onset disease accounts for 85% of neonatal GBS infection which takes place in the first week of life whereas late-onset disease occurs from the 1st week through 3 months of life.[2] Maternal GBS colonization is an important predictor of neonatal infections as vaginal transmission during delivery results in early-onset neonatal disease whereas the risk of acquiring this infection increases >25 times in neonates of colonized women in comparison to non-colonized women because 40–75% of neonates acquire GBS from the colonized mothers during passage through the birth canal.[3-5] Age, parity, education, socioeconomic status, and maternal factors such as premature rupture of membranes are the risk factors associated with maternal GBS colonization.[6] The rate of maternal GBS colonization across the world is highly varied in the literature with a mean prevalence of 17.9% and in one of the meta-analyses, the prevalence of GBS colonization among Indian pregnant women was estimated to be 7.8%.[7,8] In fact, the American College of Obstetricians and Gynecologists has recommended universal GBS screening for pregnant women at 35–37 weeks gestational age. Those mothers who test positive for GBS at 35–37 weeks of pregnancy should be considered for intrapartum antibiotics except for those having absolute medical contraindications or in cases in which cesarean section is planned with intact membranes.[9]
Madhya Pradesh among all Indian states has the highest tribal population accounting for 21.1% of the total population. The tribes mostly inhabit remote areas with less than desired healthcare facilities and are the most vulnerable section of society.[10] The fourth National Family Health Survey (India) data reported the infant mortality rate among M.P.’s tribal population to be 70% of that of its general population which means that children of one-quarter of the state’s population are 70% more likely to die, despite significant efforts of the Madhya Government to improve maternal and child health.[11] For Pradesh sustainable development, it is very important to target this vulnerable section of society and provide them with targeted healthcare services.
There is no such data in the literature providing the burden of GBS among pregnant tribal women of Madhya Pradesh. Therefore, this study was conceived and carried out at Betul, a designated tribal district of Madhya Pradesh and the Betul district hospital does not have a diagnostic facility for identification of GBS. Two primary objectives of this study were to determine the burden of GBS among 35–37 weeks pregnant females of tribal district and to explore the feasibility/effectiveness of hub-spoke model on pilot basis for collection, transport of recto-vaginal samples from Betul (spoke) to All India Institute of Medical Sciences (AIIMS) Bhopal (the hub) for performing conventional culture for GBS and polymerase chain reaction (PCR), which is not dependent on the bacterial viability. The second objective assumes much importance in the context that we were of the view that the absence of diagnostic facilities at district/peripheral healthcare settings should not be the reason for depriving the marginalized population of having access to diagnostic facilities available at a radius of 250–300 km and hub-spoke model can be effectively used by efficient operational integration. In addition, we also explored the effectiveness of diagnostic techniques such as PCR concerning conventional culture techniques in determining GBS infection.
MATERIALS AND METHODS
Study design
This was a pilot study carried out from October 2019 to March 2022 in the Department of Microbiology, AIIMS Bhopal to assess the feasibility of the hub-and-spoke model. Since this was a pilot study, we considered only a single spoke for assessing the utility of the hub-and-spoke model for GBS diagnosis in the women reporting to a healthcare facility distantly located from the tertiary care hospital hub. In this model, AIIMS Bhopal and Betul district hospitals were considered as hub and spoke, respectively, in being a tribal district located approximately 200 km from the Hub. The samples were collected at Betul district hospital (Spoke) and subsequently sent to AIIMS Bhopal (Hub) to determine GBS burden [Figure 1]. All 35–37 weeks pregnant females attending the obstetrics and gynecology department at Betul District Hospital were screened for the presence of GBS after taking informed consent. Ethical clearance was obtained from the Institutional Human Ethical Committee (LOP/2019/EF0133) before the implementation of the protocol. During the study, periodic hands-on training programs on reproductive tract infection awareness with a main emphasis on GBS, its diagnosis, treatment, and sample collection modalities were conducted for the healthcare workers by the AIIMS Bhopal investigators team. In fact, assessment of this teaching program has also been done to ensure the preparedness among the medical professionals regarding these reproductive infections. Only after training of the medical officers and nursing staff, they were asked to collect samples from eligible women.
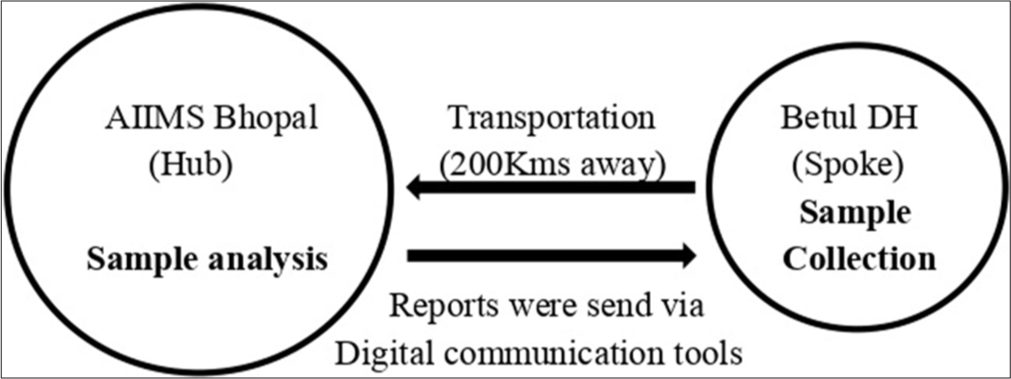
- Pictorial representation of hub-and-spoke model.
Sample collection
Recto-vaginal samples were collected from the 35 to 37 weeks pregnant women. A single-use pre-sterile HiCulture transport swab (HiMedia) was used to obtain the screening specimen first from the vagina (near the introitus) by inserting the swab about 2 cm and then from the rectum by inserting the same swab 1 cm through the anal sphincter to maximize the culture recovery which becomes important as the samples were transported from distantly location Betul district Hospital. The swab was immediately transferred into a customized container holding 7 mL transport media containing Lim broth; Todd Hewitt broth with Colistin and Nalidixic acid, at the spoke. The samples were transported to AIIMS Bhopal which was a hub maintaining cold chain for further processing.
Sample processing and culture of the specimen
After receiving the samples, the transport medium containing the Lim broth was incubated at 37°C overnight. Direct plating was carried out by inoculating swabs immersed in broth onto chromogenic agar (HiCrome Strep B Selective Agar base, HiMedia). These plates were then incubated at 37°C for 48 h under aerobic conditions as per the manufacturer’s protocol. The purple color colonies obtained were presumably considered as GBS, and confirmed by conventional microbiological methods whereas blue-green color colonies were presumably considered as Enterococcus spp, according to manufacturer’s guidelines, and further confirmed by bile-esculin test. The colonies that came out to be positive for bile-esculin were identified as Enterococcus whereas those that came out to be negative for bile-esculin were further tested for their susceptibility to bacitracin and cotrimoxazole and subsequent confirmation to GBS which is resistant to bacitracin and cotrimoxazole. Antibiotic susceptibility testing was performed using the Kirby–Bauer disc diffusion method.
DNA extraction
DNA extraction was carried out manually using the phenol-chloroform extraction method. Briefly, approximately 5 mL transport medium containing Lim broth was pelleted down. The supernatant was discarded, and the obtained pellet was resuspended in the Tris-EDTA buffer containing lysozyme (1 mg/mL) and then incubated for 30 min at 37°C. 25 µL of Proteinase K (20 mg/mL) was added and incubated at 60°C for 20 min. After this 100 µL of 10% sodium dodecyl sulfate was added and further incubated at 60°C for 20 min. 200 µL of 5M NaCl was then added and mixed well followed by the addition and mixing of 160 µL Cetyltrimethylammonium bromide and incubation at 60°C for 10 min. An equal volume of phenol-chloroform was used to separate the DNA. 0.7 volume of isopropanol was used to precipitate the DNA. Washing was performed using 70% alcohol. The obtained pellet of DNA was air-dried and resuspended in TE buffer. The yield and purity of DNA was determined using NanoDrop.
Conventional PCR
Conventional PCR was carried out to detect GBS presence using specific primers targeting atr gene responsible for the glutamine transporter protein (gbs0538) of S. agalactiae [Table 1]. This housekeeping gene and the corresponding primers were selected from the S. agalactiae Multi Locus Sequence Typing website – http://pubmlst.org/sagalactiae. 16S ribosomal ribonucleic acid (rRNA) was used as an internal control to ensure the presence of the extraction of bacterial DNA. DNA isolated from American Type Culture Collection strain (13813) of S. agalactiae was used as a template in positive control.
| Target gene | Primers | Primer sequences 5'-3' | Amplicon size |
|---|---|---|---|
| 16S rRNA | 27F | AGAGTTTGATCCTGGCTCAG | 400bp |
| 338R | GCTGCCTCCCGTAGGAGT | ||
| Atr | atrF | CGATTCTCTCAGCTTTGTTA | 779bp |
| atrR | AAGAAATCTCTTGTGCGGAT |
rRNA: Ribosomal ribonucleic acid, PCR: Polymerase chain reaction
The reaction mixture for PCR reaction included ×1 Promega buffer with a final concentration of 2.5 mM magnesium chloride, 0.25 mM of deoxynucleoside triphosphate, and 0.2 µM of each primer (forward and reverse). Approximately 30 ng of DNA was used as a template.
DNA amplification began by incubating at 94°C for 3 min followed by 35 PCR cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s. The final extension was at 72°C for 10 min for both the primer sets. After completion of PCR, gel electrophoresis was run using a 100 Bp ladder as marker to determine the desired band size.
Statistical methods
Statistical tests were performed using Microsoft Excel. Participants’ demographics are presented as mean with standard deviations. GBS-positive rate and its susceptibility to antibiotics were estimated by a proportion and summarized as a percentage. The sensitivity and specificity of PCR are determined by taking culture as a gold standard and proportions compared using exact binomial 95% confidence intervals (95% CI). P < 0.05 was considered statistically significant.
RESULTS
Maternal socio-demographics and clinical profiling
The socio-demographic and clinical profiling of the 808 pregnant females screened for the presence of GBS is summarized below in Table 2. The mean age was 24.84 ± 3.97 (range: 17– 39 years). Vaginal discharge (89.23%) was found to be the most common symptom followed by genital rashes (15.71%), fever (1.98%), leaking (1.60%), and bleeding (1.11%) at the time of presentation. About 51.23% of women have a parity of 1 or above. The most common pregnancy-associated complications found were hypertension and anemia (the majority 76.63% being in the category of mild anemia according to the WHO).
| Parameters | Value |
|---|---|
| Age (in years) | |
| <20 | 54 |
| 20–30 | 658 |
| >30 | 96 |
| Occupation | |
| Homemaker | 808 |
| Others | 0 |
| Parity index | |
| Primigravida | 389 |
| Parity 1 or above | 414 |
| Pregnancy complications | |
| Hypertension* | |
| Systolic blood pressure >140 mmHg | 14 |
| Diastolic blood pressure >90 mm Hg | 39 |
| Hemoglobin g/dL** | |
| 9–10.9 g/dL | 617 |
| 7–8.9 g/dL | 59 |
| <7 g/dL | 6 |
| Frequency of symptoms at the time of sample collection | |
| Vaginal discharge | 721 |
| Genital rashes | 127 |
| Fever | 16 |
| Bleeding | 9 |
| Leaking | 13 |
Comparative effectiveness of PCR and conventional culture method while implementing hub-and-spoke model for sample transportation
Despite Betul District Hospital being located at a considerable distance from the central hub, AIIMS Bhopal, where samples collected at Betul District Hospital were sent for analysis, a significant number of GBS-positive cases were identified using both conventional and molecular methods. This highlights the effectiveness of the hub-and-spoke model in assessing the burden of GBS, even among women residing in remote areas with limited access to facilities. In addition, we assessed the reliability of conventional culture and PCR methods for detecting GBS positivity, considering the long-distance transportation of the samples.
Table 3 shows the sensitivity and specificity of the conventional PCR assay for the detection of GBS in rectovaginal swab samples compared with the results of the conventional culture-based method.
| PCR | Culture | Total | ||
|---|---|---|---|---|
| Positive | Negative | |||
| Positive | 52 | 64 | 116 | P<0.01 with contingency correction |
| Negative | 0 | 692 | 692 | |
| Total | 52 | 756 | 808 | |
PCR: Polymerase chain reaction, GBS: Group B Streptococcus
Of the total 808 processed samples, GBS was detected in 116 (14.35%) and 52 (6.43%) by PCR and culture-based methods, respectively. All the samples in which GBS was isolated in the culture were also positive by PCR assay, therefore resulting in 100% PCR sensitivity (95%CI: 93.15– 100). Whereas of the 756 culture-negative samples for the GBS, 64 were positive with the PCR and 692 were negative for both culture and PCR indicating specificity of 91.53% (95% CI: 89.32–93.42) for the PCR, considering the culture as the gold standard.
Antibiotic susceptibility profiling of culture-positive GBS isolates
GBS isolates were found to be 100% sensitive to quinupristin – dalfopristin, linezolid, cefepime, and vancomycin, whereas 10%, 23%, 27%, and 23% of the isolates were found to be resistant to penicillin, clindamycin, erythromycin, and levofloxacin, respectively [Table 4 and Figure 2].
| Antibiotic (Potency) | Resistant isolates (%) |
|---|---|
| Penicillin (10 units) | 5 (9.6) |
| Azithromycin (15 µg) | 8 (15) |
| Levofloxacin (5 µg) | 12 (23) |
| Clindamycin (2 µg) | 12 (23) |
| Erythromycin (15 µg) | 14 (27) |
| Quinupristin – dalfopristin (15 µg) | Nil (0) |
| Linezolid (30 µg) | Nil (0) |
| Cefepime (30 µg) | Nil (0) |
| Vancomycin (30 µg) | Nil (0) |
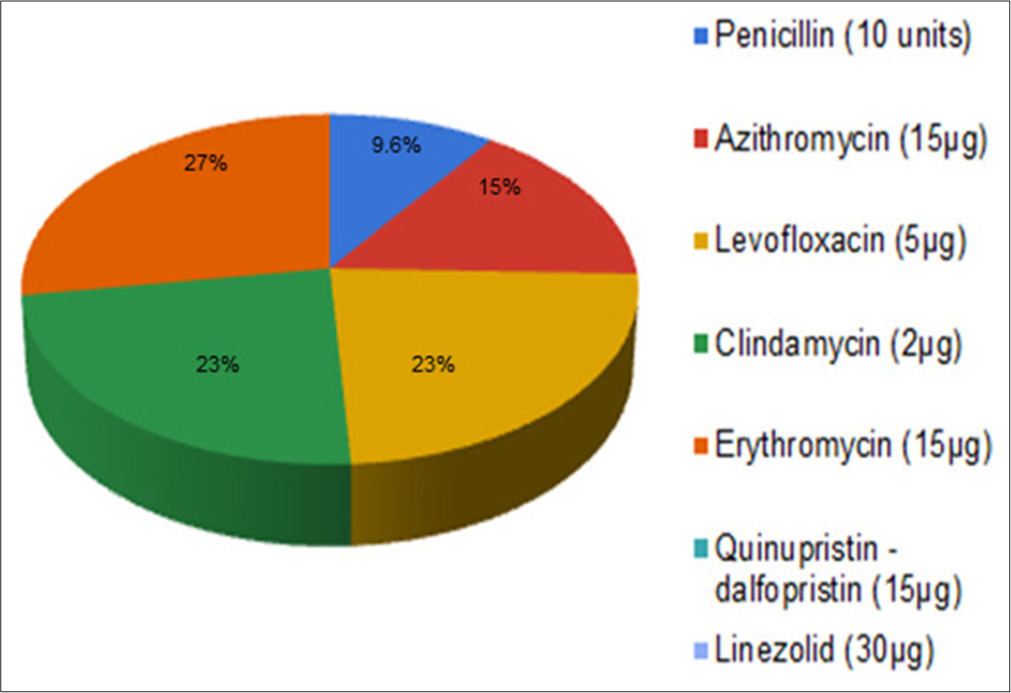
- Antibiotic susceptibility test profiling of Group B Streptococcus clinical isolates obtained through culture.
DISCUSSION
Neonatal GBS infections are significantly associated with neonates born of GBS colonized pregnant females and are also linked with high neonatal mortality rates. This is because the risk of GBS transmission increases during normal vaginal delivery in GBS colonized mothers. It has been seen that the incidence of neonatal GBS disease decreases up to 80% while implementing Centers for Disease Control and Prevention (CDC) recommended strategies such as maternal intrapartum antibiotic prophylaxis. Despite this, the burden of GBS disease remained significant leading to revised CDC guidelines in 2010 recommending universal screening modalities for diagnosing GBS colonization among all 35th–37th week pregnant females.[9,12] In one of the meta-analyses from 37 different countries, the overall mean prevalence of GBS disease was 17.9% with the highest being in Africa (22.4%), followed by America (19.7%), Europe (19.0%), South East Asia (11.1%).[7] In fact, in India, the burden of GBS colonization remained high ranging between 2% and 62% indicating the significant burden of this infection even after the issuance of revised screening guidelines by the CDC.[13] One of the reasons for the high burden of GBS colonization is the inefficient implementation of screening guidelines. Socio-economic and literacy factors, lack of awareness, and improper diagnostic facilities at peripheral hospital settings are some of the factors that become barriers to effective screening for GBS infection. This led us to explore the feasibility of a hub-and-spoke model for diagnosing GBS among 35–37 weeks pregnant women living in remote areas.
One of the studies reported a substantial increase in the culture yield when swabbing is done from both the vagina and rectum; hence, recto-vaginal swab samples were collected in our study and the prevalence of GBS was found to be 6.43% and 14.35% through conventional culture and PCR assay, respectively. Although the CDC recommended culture as the gold standard with the recent advancement in molecular methods and its appropriateness, the guidelines also included expanded laboratory methods to identify the organism. Efficient and accurate identification of an organism plays a significant role in identifying women requiring intrapartum antibiotic prophylaxis through screening. Therefore, the development of a diagnostic method with high sensitivity and specificity is of utmost importance. Moreover, in our study, the primary objective was to assess the feasibility of the hub-and-spoke model for evaluating the burden of GBS. Samples were received from the distantly located Betul district hospital, highlighting the challenges associated with transporting samples over long distances. It is well-established that with prolonged incubation of swabs in a transport medium, the viability of the organisms can decrease. Recently, both conventional and real-time PCR have largely explored other than culture methods for identification.[9] Therefore, in addition to culture, we also evaluated the GBS burden using PCR to compare the effectiveness of both methods in detecting GBS, particularly considering the challenges posed by sample transportation over long distances. In our study, we found 100% sensitivity by PCR indicating it to be much more sensitive than the conventional culture method. These results are in accordance with the other studies reporting 100% sensitivity of PCR.[14,15] Reasons for the PCR being this sensitive could be due to the usage of enrichment media before DNA extraction whereas the specificity in our study is reported to be 91.53% which also holds an importance in terms of identifying the women requiring antibiotic prophylaxis therapy. As this is definitive evidence suggestive of women to be truly negative for GBS infection, higher PCR positivity as compared to culture in all likelihood can also be ascribed to the fact that the PCR-based assay’s functions by targeting the genetic material of an organism and its viability are not essential, which is a must requirement for culture. Further, the PCR can pick up and amplify minute DNA quantity whereas for culture good numbers of viable, bacteria are required to provide culture positivity.[16] This might be one of the reasons for obtaining fewer positive cases in our study through the conventional culture technique as the samples were transported from sites approximately 200 km away. Furthermore, vaginal and perianal areas (sample collection sites) consist of lots of commensal flora which might mask or inhibit the growth of GBS. This could be another reason for missing out on the GBS colonies on culture, whereas the high sensitivity of PCR proves to be advantageous as it detects the organisms even if its genome is present in a low copy number. However, in this study, we used chromogenic selective agar for the detection of GBS which proves to be advantageous over other media as it distinguishes GBS from the other organisms based on the color. Furthermore, this agar is selective for the growth of GBS and is thought to minimize the masking effect of other common flora like E. Coli and Staphylococcus aureus over GBS.
Although the choice of drug for treating GBS colonization is penicillin, clindamycin and erythromycin are also the second choice of drugs among those allergic to penicillin[17] which showed resistance in 23% and 27% isolates, respectively, comparable to other studies. Hence, antibiotic susceptibility should be performed in women with GBS colonization but allergic to penicillin, failing which treatment failure is probable in approximately a quarter of cases. There are few isolates sensitive to erythromycin but resistant to clindamycin (presumable L phenotype), which is an uncommon phenomenon and would require further investigation. In all cases with leaking/rupture of membrane and clinical symptoms, suggestive of enhanced risk for adverse maternal-child outcome judicious usage of vancomycin may be indicated. The drawbacks of this study comprise the inability to follow up with the women and their neonates which could provide us with in-depth knowledge of the GBS transmission spectrum in utero among colonized females and assessment of multiple risk factors associated with the GBS colonization.
Apart from this, the usage of real time PCR would have been better for the identification of GBS in comparison to conventional PCR due to its advantageous sensitivity. However, this provides us the effectiveness of using conventional PCR in terms of broad spectrum settings where working with conserved resources is more feasible because any time it is far better than culture technique.
CONCLUSIONS
Hub-spoke model for GBS screening must be emphasized in rural settings to identify the pregnant women requiring prophylactic antibiotic therapy to reduce the transmission rate to neonates thereby reducing the morbidity/mortality among neonates, whereas PCR could be used as an efficient tool for identifying GBS; however, implementation of the same in the routine clinical settings needs to be analyzed for its cost-effectiveness. However, after COVID-19 diagnostic facilities are well equipped with real-time PCRs even at the district hospitals making tests for GBS identification feasible to perform. Antibiotic susceptibility profiling should be considered before prescribing antibiotics to GBS-affected women especially those allergic to penicillin.
Author contribution
SP: Conceptulized and implemented the study protocol. He formulated this manuscript in its final shape; PG: Performed experiments related to conventional culture, molecular assays and AST profiling. She has also contributed in writing this manuscript; LR: Collected samples from district hospital Betul and overall coordinated their transportation to AIIMS Bhopal; JY: Contributed in data analysis and in writing this manuscript; AH, KP, and AB: Helped in designing of inclusion and exclusion criteria, screening and recruitment of participants; AM: Helped in arranging funds for the execution of this project.
Ethical approval
The research/study was approved by the Institutional Review Board at All India Institute of Medical Sciences Bhopal, number IHEC-(LOP/2019/EF0133), dated 4th October 2019.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship: National Health Mission, MP Government, India.
References
- Group B streptococcus and pregnancy In: StatPearls. Treasure Island, FL: StatPearls Publishing; 2024. Available from: http://www.ncbi.nlm.nih.gov/books/NBK482443 [Last accessed on 2024 Aug 01]
- [Google Scholar]
- Understanding factors in Group B streptococcus late-onset disease. Infect Drug Resist. 2021;14:3207-18.
- [CrossRef] [PubMed] [Google Scholar]
- Group B Streptococcal maternal colonization and neonatal disease: Molecular mechanisms and preventative approaches. Front Pediatr. 2018;6:27.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of early-onset neonatal infection among newborns of mothers with bacterial infection or colonization: A systematic review and meta-analysis. BMC Infect Dis. 2015;15:118.
- [CrossRef] [PubMed] [Google Scholar]
- Policy statement-recommendations for the prevention of perinatal group B streptococcal (GBS) disease. Pediatrics. 2011;128:611-6.
- [CrossRef] [PubMed] [Google Scholar]
- Risk factors for group B streptococcus colonization among pregnant women in Korea. Epidemiol Health. 2011;33:e2011010.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of maternal colonisation with group B streptococcus: A systematic review and meta-analysis. Lancet Infect Dis. 2016;16:1076-84.
- [CrossRef] [PubMed] [Google Scholar]
- Meta-analysis on prevalence of vaginal group B streptococcus colonization and preterm births in India. J Matern-Fetal Neonatal Med. 2022;35:2923-31.
- [CrossRef] [PubMed] [Google Scholar]
- Prevention of Group B streptococcal early-onset disease in newborns. Available from: https://www.acog.org/clinical/clinical-guidance/committee-opinion/articles/2020/02/prevention-of-group-b-streptococcal-early-onset-disease-innewborns [Last accessed on 2024 Aug 01]
- [Google Scholar]
- Available from: https://aiggpa.mp.gov.in/uploads/project/tribal_health.pdf [Last accessed on 2024 Aug 01]
- Available from: https://dhsprogram.com/pubs/pdf/FR339/FR339.pdf [Last accessed on 2024 Aug 01]
- Prevention of perinatal group B streptococcal disease--revised guidelines from CDC, 2010. MMWR Recomm Rep Morb Mortal Wkly Rep Recomm Rep. 2010;59:1-36.
- [Google Scholar]
- Disease burden due to Group B Streptococcus in the Indian population and the need for a vaccine-a narrative review. Ther Adv Infect Dis. 2021;8:20499361211045253.
- [CrossRef] [PubMed] [Google Scholar]
- Intrapartum PCR-assay for detection of Group B Streptococci (GBS) Eur J Obstet Gynecol Reprod Biol X. 2019;4:100081.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of culture and PCR methods for diagnosis of group B streptococcus carriage in Iranian pregnant women. Iran J Public Health. 2012;41:65-70.
- [Google Scholar]
- PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4:337-48.
- [CrossRef] [PubMed] [Google Scholar]
- Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recomm Rep Morb Mortal Wkly Rep Recomm Rep. 2002;51:1-22.
- [Google Scholar]






