Translate this page into:
In silico Analysis of TCR Vβ7 of Two Patients with Type 1 Diabetes Mellitus
Address for correspondence: Dr. Jianwei Zhou, E-mail: immunolife@126.com
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective:
To compare the sequences and crystal structures of variable region of beta chain 7 (Vβ7) of T cell receptor (TCR) of two patients with type 1 diabetes mellitus (T1DM).
Patients and Methods:
The skewness of TCR Vβ7 of two T1DM patients were detected with real-time florescence quantitative polymerase chain reaction (FQ-PCR) and deoxyribonucleic acid (DNA) melting curve analysis technique followed by being sequenced, the crystal structures of them were simulated according to CPH models 2.0 Server, IMGT database, and RasMol 2 software.
Results:
The whole sequences of TCR Vβ7 of T1DM patient-1 were “CASRTAGQYEQYFGPGTR”, that of patient-2 were “CASRTAGQYEQFFGPGTR”; the only difference between them lied on the 12th amino acid. The crystal structures of Vβ7 of the two patients simulated with backbone model were rather similar, while that with sphere model were obviously different.
Conclusion:
Although the TCR Vβ7 of the T1DM patients share the similar gene sequences, their crystal structures simulated with sphere model are different, and the mechanism needs further study.
Keywords
Beta chain
crystal structure
T cell receptor
type 1 diabetes mellitus
variable region
INTRODUCTION
Type 1 diabetes mellitus (T1DM) is one of the most prevalent autoimmune diseases in the world, it is well-known that T cells play an important role in the pathogenesis of T1DM.[12345] Presently, T cell receptor (TCR) repertoire contributed to the high susceptibility of the subjects to T1DM has been reported.[6]
In the studies of Codina-Busqueta, et al.,[7] they found that the pancreatic islets were infiltrated by T cells expressing most TCR Vβ families, five of which, Vβ1, Vβ7, Vβ11, Vβ17, and Vβ22 contained monoclonal expansions as demonstrated by spectratyping and CDR3 sequencing. While Luppi, et al.,[8] reported that an increased frequency of circulating Vβ1, Vβ7, and Vβ17 T cells in peripheral blood mononuclear cells (PBMCs) from recently diagnosed T1DM patients, as compared with control subjects. Similarly, an increase in Vβ7 expression in PBMCs from T1DM patients was associated with a superantigen-mediated T cell expansion.[9] In another study on two children with T1DM, a marked overrepresentation of messenger ribonucleic acid (mRNA) encoding TCR Vβ7 chain was observed.[10] Obviously, a conclusion can be drawn from the above studies that Vβ7 was the skewed gene family with high frequency for T1DM patients.
In this study, we used real-time florescence quantitative polymerase chain reaction (FQ-PCR) and deoxyribonucleic acid (DNA) melting curve analysis technique[1112] to detect the skewness of TCR Vβ7 of two patients with T1DM, moreover, in silico analysis of the crystal structures of the two gene families was performed, and found that even if the two TCR Vβ7 of different T1DM patients contains similar sequences, their crystal structures are not coincident with each other.
PATIENTS AND METHODS
Patients
Peripheral blood lymphocytes (PBL) were collected from two T1DM patients, who were not treated with immunomodulating drugs in the previous 6 months prior to the study and were seronegative for markers of hepatitis viruses, human immunodeficiency virus (HIV), and other pathogenic infections. Excluded from the study were patients with tumors and immunological disorders. This study protocol was approved by the Hospital Ethics Committee.
Extraction of RNAS and systhesis of the first CDNA
The sense primer, antisense primer, and specific primers for Vβ7 genes were previously described[10] and synthesized by the Guangzhou Daangene Corporation of China. A total of 5 ml of blood were taken from each T1DM patients. PBL were isolated by Ficoll-Hypaque density centrifugation. Using an Omega RNA extraction kit according to the manufacturer's instructions, total RNA was extracted and 1 μg total RNA was reverse transcribed with 250 pm olig (dT), 200 U Moloney murine leukemia virus (M-MuLV) reverse transcriptase, and 2 ml of 10 mM deoxyribonucleotide triphosphate (dNTP) mix (cDNA Synthesis Kit; MBI-Fermentas), in a total volume of 20 μl (six reactions for every sample). The cDNA was stored at −80°C.
Amplification of TCR Vβ7 complementary DNA
The FQ-PCR for each Vβ7 was carried out in a 20 μl volume with 10 ml 2 × real-time PCR Master Mix (TOYOBO, JAPAN), which contained Taq-polymerase, dNTPs, PCR buffers, and SYBR green I. The final concentration of each primer was 0.3 μM, and at last, 1 μl liquid containing 10 ~ 50 ng reverse-transcribed total RNA was added to the reaction mixture as the PCR template. Reactions were performed in MJ Opticon 2 DNA engine and analyzed with Opticon Monitor 3.0 software (Bio-rad, USA), under the following process: Preincubation at 94°C for 3 min, 94°C melting for 20 s, primer annealing at 56°C for 30 s, and extension at 72°C for 30 s. The above procedure is iterated 40 cycles for the whole amplification.
Detection of TCR Vβ7 sequences
According to the melting curve, the PCR products of TCR Vβ7 were selected and again performed PCR according to the procedures of ‘AMPLIFICATION OF TCR Vβ7 CDNA’ section, then sequenced them on an ABI 377 DNA sequencer (The Corporation of Invitrogen, Shang-Hai, China).
In silico analysis of TCR Vβ7
In light of the sequences, the crystal structures of TCR Vβ7 were mutilated with IMGT database, CPH models 2.0 Server and RasMol 2 software.
RESULTS
The whole sequences of TCR Vβ7 of T1DM patient-1 were “CASRTAGQYEQYFGPGTR”, that of patient-2 were “CASRTAGQYEQFFGPGTR”;the only difference between the two patients was the 12th amino acid was “Y” in the first patient, that, while which was “F” in the second one. “TAGQYEQ” were the common specific gene sequences in TCR Vβ7 of the two T1DM patients [Table 1].

According to IMGT database, CPH models 2.0 Server, and RasMol 2 software; the crystal structures of TCR Vβ7 of the two T1DM patients [Figures 1 and 2] were simulated. In Figure 1, the crystal structures (made in backbone model) of Vβ7 of patient-1 was very similar to that of patient-2, while in Figure 2, the crystal structures (made in sphere model) of Vβ7 of the two patients looked different. According to the outlines of the three grooves and CDRs, the predicated antigen peptides of the Vβ7 of two T1DM patients were drawn out, respectively. Obviously, they were not identical with each other [Figure 3].
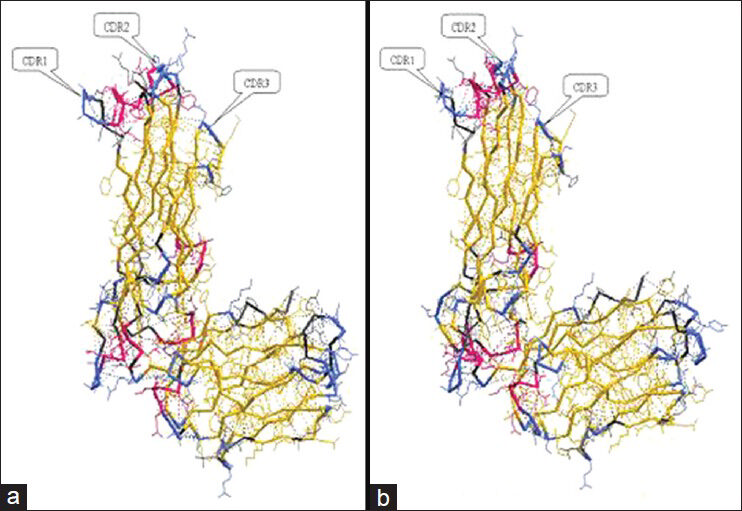
- The crystal structures (in backbone model) of T cell receptor of variable region of beta chain 7 (TCR Vβ7) of two type 1 diabetes mellitus (T1DM) patients made by CPH models 2.0 Server and IMGT database. (a) The crystal structure of TCR Vβ7 of patient-1 with T1DM. (b) The crystal structure of TCR Vβ7 of patient-2 with T1DM

- The crystal structures (in sphere model) of TCR Vβ7 of two T1DM patients made by CPH models 2.0 Server and RasMol 2 software. (a) The crystal structure of TCR Vβ7 of patient-1 with T1DM. (b) The crystal structure of TCR Vβ7 of patient-2 with T1DM

- The predicted antigen peptides specific to T1DM according to crystal structures of TCR Vβ7 of two T1DM patients. (a) The predicted antigen peptides specific to T1DM according to crystal structures of TCR Vβ7 of patient-1 with T1DM. (b) The predicted antigen peptides specific to T1DM according to crystal structures of TCR Vβ7 of patient-2 with T1DM
DISCUSSION
TCR Vβ7 gene families overexpressed in PBL of the patients with T1DM, and the skewness of Vβ7 was thought as critical features specific to T1DM.[78910] In this study, Vβ7 of the two T1DM patients were still the biased genes which shared the same sequences “TAGQYEQ”; in some reports,[71113] these common sequences were called gene melting spectral patterns (GMSPs). In the study by Yang, et al.,[14] the T cells which TCR Vβ contains such GMSPs were taken as the specific T-cell clone to the immunogen. Many researchers reported that only the clonal proliferative T cells play important roles in the process of recognizing and removing antigens from bacteria, viruses, and tumors.[111215] According to the results of this study, the authors also thought that TCR Vβ7 is the key clonally proliferational gene specific to T1DM, and its bias is probably one of the factors for onset or development of T1DM.
With IMGT database, CPH models 2.0 Server and RasMol 2 software, we simulated the crystal structure of TCR Vβ7 of the two T1DM patients. With the backbone model [Figure 1], due to the similarities between the sequences of the two Vβ7 genes, the two pictures of them are so similar that it is hard to check out the difference between them. Interestingly, with the sphere model [Figure 2], it is easy to found out that the difference between the structure of Vβ7 of patient-1 and that of patient-2. Further, drawing a line on the CDRs and grooves of the two crystal structures respectively, the two lines were obviously different, because they could hardly overlap as shown in Figure 3. To some degree, the lines are equal to the antigen peptide-major histocompatibility complex (MHC) specific to T1DM, because they are the complementary and combining positions for CDRs and peptide-MHC. Therefore, for most of T1DM patients, it is possible that the skewness of Vβ7 is alike, but the causal immunogens specific to T1DM are probably different. While in the previous studies,[11121316] the researchers found the common GMSPs in different patients with a certain disease and considered that this may be caused by the same antigen epitopes; however, according to this study, the argument probably needs further discussion.
CONCLUSION
Vβ7 is the common biased gene family for the two T1DM patents, although there are very similar sequences, their crystal structures simulated with sphere model exhibit obviously different. This not only indicates the same skewness of TCR Vβ is caused by different antigen peptides in certain a disease, but also proves that in silico analysis technique such as simulation with sphere model could be used as a compliment method for detecting skweness of TCR Vβ. However, due to the cases for this experiment is small, the arguments need more studies to further prove in future.
ACKNOWLEDGEMENTS
All the authors thank the two T1DM patients for providing the research samples, and the Science Research Department of Jining Medical College and of the Affiliated Hospital for supporting this study. Subsequently, we also thank the grant from the Provincial Science and Technology Development Project (No. 2012YD18054), the Provincial Nature Science Foundation (No. ZR2012HL29), the High School Science and Technology Plan Project (No. J11LF18), the Population and Family Planning Commission (No. [2011]13), the Development Plan Project of Jining Science and Technology Bureau of Shandong Province (No. [2011]57), and the Youth Foundation of Jining Medical College (No. [2011]).
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- B7-2(CD86) controls the priming of autoreactive CD4 T cell response against pancreatic islets. J Immunol. 2004;173:3631-9.
- [Google Scholar]
- Thymectomy and radiation-induced type 1 diabetes in nonlymphopenic BB rats. Diabetes. 2002;51:2975-81.
- [Google Scholar]
- Impaired post-thymic development of regulatory CD4+CD25+T cells contributes to diabetes pathogenesis in BB rats. J Immunol. 2005;174:4081-9.
- [Google Scholar]
- Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol. 2009;155:173-81.
- [Google Scholar]
- Gene expression profiles for the human pancreas and purified islets in type 1diabetes: New findings at clinical onset and in long-standing diabetes. Clin Exp Immunol. 2010;159:23-44.
- [Google Scholar]
- Analysis of the rat Iddm14 diabetes susceptibility locus in multiple rat strains: Identification of a susceptibility haplotype in the Tcrb-V locus. Mamm Genome. 2009;20:162-9.
- [Google Scholar]
- TCR bias of in vivo expanded T cells in pancreatic islets and spleen at the onset in human type 1 diabetes. J Immunol. 2011;186:3787-97.
- [Google Scholar]
- Restricted TCR V beta gene expression and enterovirus infection in type I diatetes: A pilot study. Diabetologia. 2000;43:1484-97.
- [Google Scholar]
- Evidence for superantigen involvement in insulin-dependent diabetes mellitus aetiology. Nature. 1994;371:351-5.
- [Google Scholar]
- T cell activation by coxsackievirus B4 antigens in type 1 diabetes mellitus: Evidence for selective TCR Vβ usage without superantigenic activity. J Immunol. 2001;167:3513-20.
- [Google Scholar]
- Primary exploration of the third complementary determining region spectratyping and molecular features of T cell receptor alpha chain in the peripheral blood and tissue of patients with colorectal carcinoma. ACTA Medica Mediterranea. 2011;27:23-30.
- [Google Scholar]
- Comparing TCR beta chain variable gene skewness between children with tuberculosis and BCG-vaccinated children. Arch Iran Med. 2013;16:104-8.
- [Google Scholar]
- Primary exploration of molecular and spectratyping features of CDR3 of TCR β chain in the peripheral blood and tissue of patients with colorectal carcinoma. Cancer Epidemiol. 2010;34:733-40.
- [Google Scholar]
- Rapid detection of clonal expansion of T-cell receptor-beta gene in patients with HBV using the real-time PCR with DNA melting curve analysis. Hepatol Res. 2010;40:407-14.
- [Google Scholar]
- T cell receptor diversity of CD8+T lymphocytes and its association with viral load in individuals with HIV-1 infection. Zhonghua Yu Fang Yi Xue Za Zhi. 2009;43:404-8.
- [Google Scholar]
- Analysis of the CDR3 region of alpha/betaT-cell receptors (TCRs) and TCR BD gene double-stranded recombination signal sequence breaks end in peripheral blood mononuclear cells of T-lineage acute lymphoblastic leukemia. Clin Lab Haematol. 2006;28:405-15.
- [Google Scholar]





