Translate this page into:
Detection and genetic characterization of extended-spectrum beta-lactamases producers in a tertiary care hospital
Address for correspondence: Dr. Sarita Otta, 1/16, Kanchanjunga Enclaves, Chandrasekharpur, Bhubaneswar, Odisha, India. E-mail: saritaotta@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Extended-spectrum beta-lactamase (ESBL)-producing organisms inactivate extended beta-lactam antibiotics and monobactams and also exhibit coresistance to many other classes of antibiotics. The present study was carried out to assess the prevalence of the ESBLs and to determine the most prevalent genotype in our hospital.
MATERIALS AND METHODS:
All clinically significant Gram-negative isolates were identified, and their antimicrobial susceptibility testing was done by Kirby–Bauers' disc diffusion method. ESBL detection was confirmed by minimal inhibitory concentration method using agar dilution technique for those who screened positive by ceftazidime (30 μg) disc. Further, the established ESBL-positive isolates were subjected to genotyping for bla TEM, bla CTX-M, and bla SHV genes by using conventional polymerase chain reaction.
RESULTS:
Escherichia coli was the most common (28.84%) Gram-negative bacillus followed by Klebsiella pneumoniae (18.07%), while Pseudomonas spp. (9.61%) was the most commonly identified nonfermenter. ESBL production was detected in 160 (30.8%) isolates. Klebsiella oxytoca (46.7%) followed by E. coli (44%) were the common ESBL producers. Most predominant ESBL gene was bla TEM, found in 122 (76.25%) isolates. Combinations of two genes were seen in 109 (68.1%) isolates, the most common (43.12%) combination being blaTEM and blaCTX-M. In this study, 16 (10%) strains had all the three types of genes. Most of the isolated Gram-negative bacilli (GNB) were sensitive to amikacin, imipenem, and colistin.
CONCLUSION:
In our study, the 30.8% of GNB were ESBL producers. This is the only study that shows that TEM is the most prevalent ESBL genotypes in our area. Of concern is a good number of isolates showing all three patterns of genes (TEM, SHV, and CTX-M). Amikacin, imipenem, and colistin were the most useful antibiotics in our setup.
Keywords
Bla TEM
extended-spectrum beta-lactamase
genotyping
Introduction
With the broadening front of microorganisms having enzymes capable of destroying beta-lactam drugs, a newer class of drugs known as oxyimino-cephalosporins were developed for the treatment of serious infections due to Gram-negative bacteria to which the microbes responded by creating an enzyme capable of hydrolyzing them. These enzymes named as extended-spectrum beta-lactamases (ESBLs) have a propensity to inactivate extended-spectrum beta-lactam antibiotics and monobactams except cephamycins and imipenem. In addition, ESBL-producing organisms exhibit coresistance to many other classes of antibiotics resulting in limitation of therapeutic option. These are Ambler Class A β-lactamases, commonly having genotypes of such as bla SHV, TEM, and CTX-M types. The present study was undertaken to note the prevalence of ESBL Gram-negative bacilli (GNB) as well as the predominant genotypic pattern.
Materials and Methods
This prospective study was conducted in our department after approval by the institutional ethical committee. During the study period of 2 years, 1509 clinical samples were collected from indoor, outdoor, as well as intensive care unit (ICU) patients. The samples were processed, and the cultures were identified as per the standard protocol. Antibiotic susceptibility testing of all the isolates was carried out by Kirby–Bauer's disc diffusion method and interpreted according to the CLSI guidelines. All the GNB were screened for ESBL production by using ceftazidime (CAZ) (30 μg) discs. The organisms which showed a zone diameter <22 mm were subjected to further confirmatory tests.
Minimum inhibitory concentration (MIC) was carried out by agar dilution technique against antibiotics CAZ and ceftazidime-clavulanic acid (CAC) as per the CLSI breakpoints. About 0.0625 to 32 μg/ml concentrations of CAZ was obtained, and for CAC, 4 μg/ml of clavulanate were added to the respective CAZ dilutions. The lowest concentration of antibiotic that prevented bacterial growth was considered to be the MIC. Organism was considered as ESBL producer if MIC with CAZ is ≥2 μg/ml. This was further confirmed when MIC of CAC showed a reduction of ≥3 twofold dilution than that of CAZ alone[1] [Figure 1].
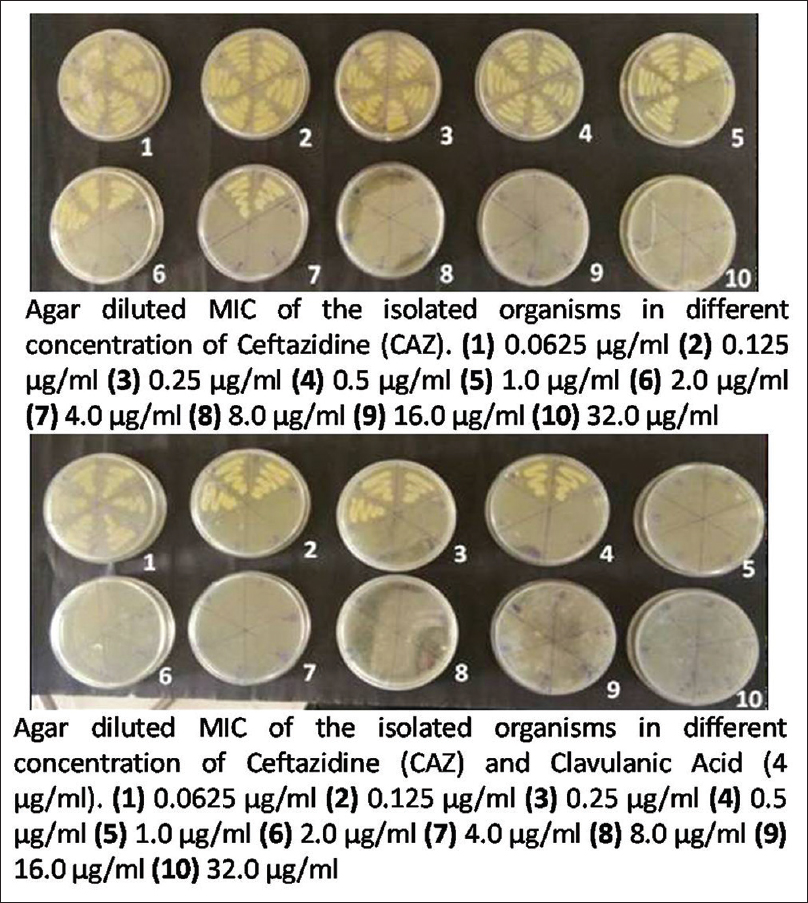
- Detection of minimum inhibitory concentration by agar dilution method using ceftazidime alone and ceftazidime with clavulanic acid
Genomic DNA was isolated from phenotypically confirmed ESBL-producing bacterial cells by lysis method[2] Extraction has been done by using bacterial genomic DNA extraction kit (HiMedia, Mumbai). The DNA samples were run on 0.8% agarose gel, stained with ethidium bromide, and examined under Gel Doc (Biorad) against uncut Lambda DNA to confirm the presence and note the quantity of DNA.
Extracted DNA was taken for polymerase chain reaction (PCR) assay to ensure the presence of the blaSHV, blaTEM, and blaCTX-M genes. About 2.5 μl of PCR buffer (without MgCl2), 2.5 μl of 25 mM MgCl2, 0.2 μl of 200 mM deoxyribonucleotide triphosphate, 1 μl of each forward and reverse primers[34](10–12 pmole) [Table 1], and 0.1 μl of 5U Taq polymerase (HiMedia) were used for a single reaction and 1 μl of the extracted genomic DNA was added to the reaction mixture. The volume was made up to 25 μl by using deionized water.
| Primer name | Primer sequence | Size of the PCR product (bp) | Reference |
|---|---|---|---|
| TEM (bla TEM) | F 5’- AAGCCATACCAAACGACGAG-3’ | 108 bp | Yano et al., 1999[3] |
| R 5’- ATTGTTGCCGGGAAGCTAGA-3’ | |||
| SHV (bla SHV) | F 5’- TCTCCCTGTTAGCCACCCTG-3’ | 593 bp | Yano et al., 1999[3] |
| R 5’- CCACTGCAGCAGCTGC (A/C) GTT-3’ | |||
| CTxM (bla CTxM) | F 5’- CGATGTGCAGTACCAGTAA-3’ | 585 bp | Batchelor et al., 2005[4] |
| R 5’- TTAGTGACCAGAATCAGCGG-3’ |
PCR=Polymerase chain reaction
The PCR machine was programmed as per the protocol in Table 2. The amplified PCR products were analyzed against 100 bp ladder (HiMedia) using 1.5% agarose gel electrophoresis. Gels were stained with ethidium bromide and visualized by ultraviolet transilluminator or Gel Doc instrument [Figure 2].
| Stage | Step | TEM | SHV | CTX-M |
|---|---|---|---|---|
| Initial denaturation | Initial denaturation | 94°C-2 min | 94°C-2 min | 94°C- 2 min |
| 1 cycle | 1 cycle | 1 cycle | ||
| Amplification | Denaturation | 94° C-1 min | 94° C-1 min | 94° C-1 min |
| 30 cycles | 30 cycles | 30 cycles | ||
| Annealing | 58° C-1 min | 52° C-30 s | 55° C-1 min | |
| 30 cycles | 30 cycles | 30 cycles | ||
| Extension | 72° C-1 min | 72°C-45 s | 72° C-1 min | |
| 30 cycles | 30 cycles | 30 cycles | ||
| Final extension | Extension | 72° C-7 min | 72° C-5 min | 72° C-5 min |
| 1 cycle | 1 cycle | 1 cycle |
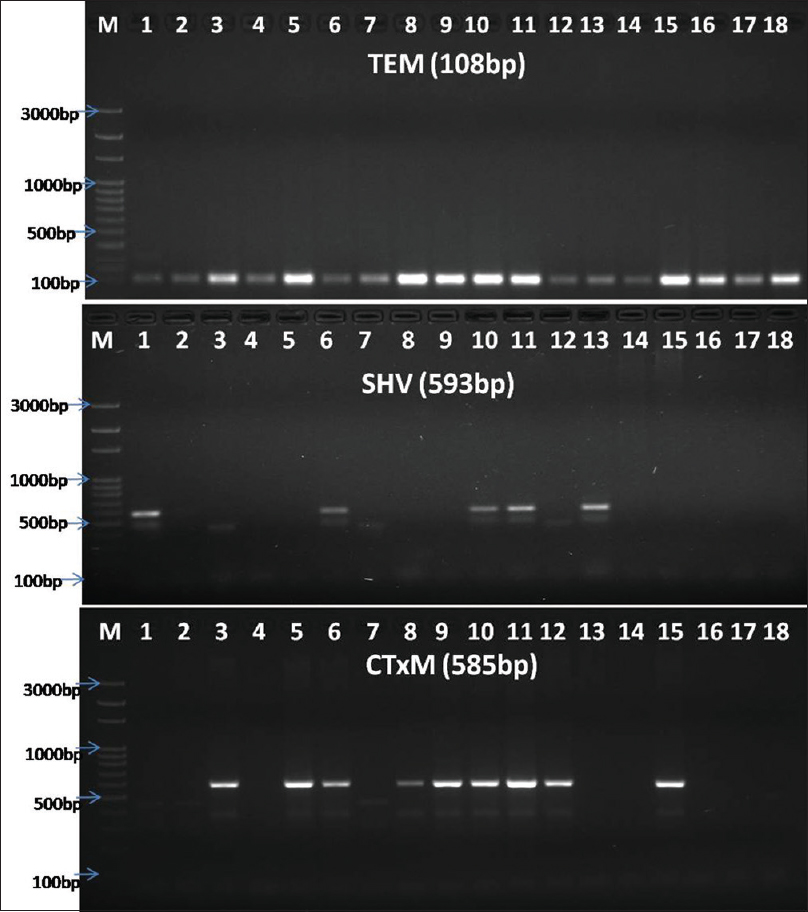
- Banding pattern of TEM, SHV, and CTX-M genes through gel electrophoresis (Lane-M: 100 bp ladder, Himedia, India, and Lane-2–18: sample. TEM positive for samples 1–18; CTX-M positive for 1, 6, 10, 11, 13; SHV positive in samples 3, 5, 6, 8, 9, 10, 11, 12, 15)
Results
A total of 1509 clinical specimens were obtained during the study period, of which 712 (47.18%) showed growth of various organisms, 520 (73.03%) yielding single Gram-negative bacteria only. Of these 520 GNBs, 309 (59.42%) were from male patients, while 211 (40.57%) from females with a male-to-female ratio of 1.46:1. Most commonly, 161 (30.96%) of GNBs were isolated from patients in the age group of 16–30 followed by 31–45 years (23.46%). Highest proportion of ESBLs was found in the samples in the age group of 46–60 years (30.62%).
Most of the samples, 418 (80.38%), were collected from the inpatient department, of which 109 (21%) cases were from ICUs. There were only 102 (19.61%) samples from the outpatient department. Most of the ESBL strains (33.9%) were obtained from samples from wards and cabins. ICUs although thought to be the harborer of resistant strains, we had only 30.3% ESBL producers.
Urine was the most predominant (54.07%) sample received during the study period. However, highest amount of ESBL producers were isolated from pus samples (60.34%) followed by those from the blood (58.06%) [Table 3].
| Samples | Total number of samples | Number of samples with GNB isolated | Number of ESBL positive (percentage of ESBL) |
|---|---|---|---|
| Blood | 182 | 31 | 18 (58.06) |
| CSF | 23 | 02 | 0 (0) |
| High vaginal swab | 15 | 03 | 0 (0) |
| Pus | 148 | 58 | 35 (60.34) |
| Sputum | 71 | 36 | 13 (36.1) |
| Stool | 6 | 2 | 0 (0) |
| Tracheal aspirate | 104 | 62 | 14 (22.58) |
| Urine | 816 | 235 | 57 (24.26) |
| Wound swab | 144 | 91 | 23 (25.27) |
| Total | 1509 | 520 | 160 |
ESBL=Extended-spectrum beta-lactamases, GNB=Gram-negative bacilli, CSF=Cerebrospinal fluid
Escherichia coli were the most common GNB (28.84%) isolated followed by Klebsiella pneumoniae (18.07%) and Citrobacter freundii (7.7%). Pseudomonas spp. (9.61%) followed by Acinetobacter spp. (6.9%) were the commonly identified nonfermenters.
Of the 520 GNB, 160 isolates were ESBL producers. Klebsiella oxytoca (46.7%) were the most predominant ESBL positive organisms followed by E. coli (44%). All the isolates belonging to members of tribe Proteae, Serratia spp., Chromobacterium spp., and Shigella spp. were non-ESBL producers. Of note was Enterobacter spp. and Citrobacter spp. which were ESBL positive in 48% and 14.7% of cases, respectively. On the other hand, nonfermenters in our study were ESBL producers in 36.1% and 20% for Acinetobacter spp. and Pseudomonas spp., respectively [Table 4].
| Organisms | Total | ESBL strains (percentage of total) |
|---|---|---|
| E. coli | 150 | 66 (44.0) |
| K. pneumonia | 94 | 40 (42.6) |
| A. baumannii | 36 | 13 (36.1) |
| K. oxytoca | 15 | 7 (46.7) |
| P. aeruginosa | 50 | 10 (20) |
| E. cloacae | 14 | 8 (57.14) |
| C. koseri | 35 | 6 (17.14) |
| C. freundii | 40 | 5 (12.5) |
| E. aerogenes | 11 | 4 (36.4) |
| Salmonella Spp. | 8 | 1 (12.5) |
| P. mirabilis | 24 | 0 (0) |
| P. vulgaris | 26 | 0 (0) |
| S. marcescens | 3 | 0 (0) |
| Chromobacterium violaceum | 1 | 0 (0) |
| Providencia Spp. | 2 | 0 (0) |
| Morganella Spp. | 3 | 0 (0) |
| Shigella Spp. | 8 | 0 (0) |
| Total | 520 | 160 |
E. coli=Escherichia coli, K. pneumonia=Klebsiella pneumoniae, A. baumannii=Acinetobacter baumannii, K. oxytoca=Klebsiella oxytoca, P. aeruginosa=Pseudomonas aeruginosa, E. cloacae=Enterobacter cloacae, C. koseri=Citrobacter koseri, C. freundii=Citrobacter freundii, E. aerogenes=Enterobacter aerogenes, P. mirabilis=Proteus mirabilis, P. vulgaris=Proteus vulgaris, S. marcescens=Serratia marcescens, ESBL=Extended-spectrum beta-lactamases
In the present study [Table-5], the isolated GNB are mostly sensitive to amikacin (93.7%), imipenem (96.5%), and colistin (98.85%). ESBL producers showed a higher degree of resistance than non-ESBL producers. Amoxiclav, ofloxacin, ceftriaxone, cotrimoxazole, and imipenem activity is reduced in GNBs which show ESBL production. Other antibiotics such as cefoperazone-sulbactam, colistin, and gentamicin are the choicest antibiotics because of their higher or similar efficacy in ESBL producers than their counterparts. Nitrofurantoin showed high sensitivity in urine isolates. It is very effective in ESBL-producing organisms although less than their ESBL-negative counterparts.
| Drugs | Number of resistant organism among ESBL-producing isolates (percentage resistance) (n=160), n (%) | Number of resistant organism among non-ESBL-producing isolates (percentage resistance) (n=360), n (%) | Overall percentage of resistance (n=520), n (%) |
|---|---|---|---|
| Ampicillin (10 µg) | 157 (98.12) | 358 (99.44) | 99.03 |
| Amoxiclav (20/10 µg) | 48 (30) | 74 (20.55) | 23.46 |
| Amikacin (10 µg) | 7 (4.37) | 26 (7.22) | 6.34 |
| Ceftriaxone (30 µg) | 160 (100) | 159 (44.16) | 61.34 |
| Cefoperazone-sulbactam (75/10 µg) | 0 | 263 (73.05) | 50.57 |
| Cotrimoxazole (25 µg) | 122 (76.25) | 238 (66.11) | 69.23 |
| Cefotaxime (30 µg) | 157 (98.12) | 143 (67.50) | 57.69 |
| Gentamicin (10 µg) | 60 (37.5) | 242 (67.22) | 58.07 |
| Ofloxacin (5 µg) | 160 (100) | 285 (79.16) | 85.57 |
| Nitrofurantoin (300 µg)* | 6 (10.62) | 11 (6.18) | 7.6 |
| Imipenem (10 µg) | 15 (9.37) | 3 (0.83) | 3.46 |
| Tigecycline (15 µg) | 10 (6.25) | 40 (11.11) | 9.61 |
| Colistin (10 µg) | 1 (0.62) | 5 (1.39) | 1.15 |
*Nitrofurantoin used only in urine samples. ESBL=Extended-spectrum beta-lactamases
All the ESBL-producing isolates confirmed by phenotypic methods were analyzed for the presence of blaCTXM, blaTEM, and blaSHV genes [Table-6]. All 160 (100%) isolates showed the presence of bands in any of the three ESBL genes. Most predominant gene was TEM, found in 122 (76.25%) isolates followed by CTX-M in 115 (71.87%) and SHV in 64 (40%). Single type of ESBL gene was seen in 35 (21.9%) isolates; 109 (68.1%) had a combination of 2 genes, while only 16 (10%) strains had all the 3 types of genes. The prevalence of only blaTEM (10.62%) gene was highest followed by blaCTX-M (6.25%) and blaSHV (5%). Coexistence of two genes was observed highest with blaTEM and blaCTX-M gene in 69 (43.12%) isolates followed by blaTEM and blaSHV in 20 (12.5%) isolates and blaCTX-M and blaSHV in 20 (12.5%) isolates only. The most common genotype found in ESBL producers was coexistence of blaCTX-M and blaTEM genes accounting for 43.1% of cases.
| Organisms | ESBL number | TEM | CTxM | SHV | TEM+CTxM+SHV | TEM+ CTxM | TEM+ SHV | CTxM+ SHV |
|---|---|---|---|---|---|---|---|---|
| E. coli | 66 | 6 | 2 | 1 | 3 | 39 | 8 | 7 |
| K. pneumoniae | 40 | 4 | 2 | 2 | 8 | 15 | 4 | 5 |
| Acinetobacter spp. | 13 | 1 | 2 | 1 | 2 | 1 | 3 | 3 |
| K. oxytoca | 7 | 1 | 1 | 1 | 0 | 2 | 1 | 1 |
| Pseudomanas spp. | 10 | 0 | 0 | 1 | 2 | 2 | 2 | 3 |
| E. cloacae | 8 | 0 | 1 | 1 | 1 | 4 | 1 | 0 |
| C. freundii | 5 | 1 | 1 | 0 | 0 | 2 | 0 | 1 |
| C. koseri | 6 | 2 | 0 | 0 | 0 | 3 | 1 | 0 |
| E. aerogenes | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| Salmonella spp. | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 160 | 17 | 10 | 8 | 16 | 69 | 20 | 20 |
GNB=Gram-negative bacilli, ESBL=Extended-spectrum beta-lactamases, E. coli=Escherichia coli, K. pneumonia=Klebsiella pneumoniae, K. oxytoca=Klebsiella oxytoca, E. cloacae=Enterobacter cloacae, C. freundii=Citrobacter freundii, C. koseri=Citrobacter koseri, E. aerogenes=Enterobacter aerogenes
Discussion
ESBL-producing GNB have emerged as serious pathogens, as they are capable of hydrolyzing extended-spectrum beta-lactam antibiotics and monobactams. In addition, they exhibit coresistance to many other classes of antibiotics, thus limiting the therapeutic options. ESBLs are Ambler Class A β-lactamases commonly having genotypes bla SHV, bla TEM, and bla CTX-M. Other genotypes include bla KPC, VEB, PER, BEL-1, BES-1, SFO-1, TLA, and bla BIC.[5]
In the present study, 520 Gram-negative bacteria were isolated, of which 160 (30.8%) were ESBL producers. Prevalence of ESBL varies worldwide. A strikingly high frequency rate of ESBLs (44.9%) has been reported from Latin America,[6] whereas frequency rates in Greece, Portugal, Germany, and the Netherlands is 27.4%, 15.5%, 2.6%, and 2%, respectively.[7] ESBL prevalence in pathogens tested as part of the Surveillance of Antimicrobial Resistance (SARI) in (2004–2006) study in India was 88%.[8] Such high prevalence is probably due to rampant and inadvertent use of the third-generation cephalosporins. Over-the-counter availability could be another cause when patients resort to self-medication.[9] A previous study from our institute reported 51.78% of uropathogens as ESBL producers.[10] A reduction in the percentage of ESBL producers in our hospital is probably due to a leap in training and adherence to infection control procedures in recent past.
Maximum GNB samples in this study were isolated from wards and cabins 221 (42.5%), followed by ICUs 109 (20.96%) and Outpatient department (OPD) 102 (19.61%). Wards had the highest prevalence of ESBL-producing GNBs followed by ICUs. OPD samples were no less behind in harboring ESBL-producing GNBs. Vaguely, this suggests that in our area, the prevalence of ESBL in community is high. Although the ESBL-related prevalence studies are traditionally associated with health-care settings, there is an increasing number of reports documenting the isolation of ESBLs in community settings as well.[11]
E. coli (150) and Klebsiella spp. (109) were the two most common isolates in the present study. E. coli (44%) was the most predominant ESBL producer followed by Klebsiella spp. (43.1%). A previous study[12] ESBL production was observed in 41% of E. coli and 40% of K. pneumoniae.
Other members of Enterobacteriaceae when pondered into Enterobacter spp. showed a very high degree (48%) of ESBL production. None of the strains of Proteus spp. and Shigella spp. produced any ESBL. Nonfermenters such as Pseudomonas spp. and Acinetobacter spp. were positive for ESBL in 20% and 36% cases, respectively. Although the data for GNBs other than E. coli and Klebsiella spp. are limited, a study[13] has found 15% of Pseudomonas spp. to be ESBL producers. In an Indian study,[14]28% of the Acinetobacter spp. isolates were ESBL producing. However, these are notorious because of the transferable nature of the ESBL plasmids in Acinetobacter[15] which is further facilitated by the ability of this bacteria to survive for a long time in the hospital environment.[14] Routine detection of ESBL-producing strains may be difficult because the synergy between cephalosporin and clavulanic acid, typically observed with ESBL-producing members of Enterobacteriaceae, tends to be minimal with Acinetobacter spp.[16] Thus, our study, due to the inherent limitations, may have underestimated the prevalence of ESBL in Acinetobacter spp.
The antibiotic susceptibility testing results of all 520 isolates showed most of the isolates (72.69%) in the present study were multidrug-resistant (MDR) (resistance to three or more group of antibiotics). Similarly, higher rate of MDR was also reported by Ansari et al.[17] (78%). Amikacin, colistin, tigecycline, and imipenem were the most effective antibiotics in ESBL-producing organisms in the present study. Other studies[1819] denoted amikacin and imipenem are the choicest drugs for ESBL-positive strains. In our study, quinolones and beta-lactams showed the lowest efficacy similar to other studies.[1819] Among aminoglycosides, amikacin was a more effective antibiotic than gentamicin. This has also been noted in a similar to study by Rahman et al.[20] In our study, ESBL-positive isolates showed higher degree of resistance to amoxiclav, cotrimoxazole, ofloxacin, nitrofurantoin, beta-lactam drugs, and imipenem than the non-ESBL strains. A similar study[21] showed gentamicin, amikacin, norfloxacin, levofloxacin, ciprofloxacin, and netilmicin having a significant reduced activity in ESBL producers than the non-ESBL producers. The non-ESBL strains were more resistant to aminoglycosides than ESBL strains unlike the above study findings, thus proposing other mechanisms of aminoglycoside resistance.
There is a paucity of literature as to the genotypic pattern of ESBL producers in our area. Genotyping of 160 ESBL isolates showed that TEM 122 (76.25%) is the predominant ESBL gene in GNB isolates. Few of the recent studies corroborate this data.[922] Until the year 2000, TEM was the most prevalent ESBL gene in the Indian bacterial population but was replaced by CTX-M in the following decade.[23] Reports from South America, Israel, Spain, New York, the United Kingdom, and several parts of the Indian subcontinent have revealed CTX-M as the predominant gene.[23] In this study, CTX-M was seen in 115 (71.87%) cases either singly or in combination with other genes. CTX-M gene constitutes a distinct phylogenetic lineage of molecular class A beta-lactamases that exhibit a higher preference for cefotaxime and ceftriaxone, than CAZ, and a higher susceptibility to tazobactam than to clavulanate. They still exist among the Indian bacterial population, especially in our settings and hence are a matter of concern for clinicians.[24]
Coexistence of all the three bla genes was observed in 16 (10%) isolates in our study, which was higher than the report from North India where they observed only in 6.45% isolates.[22] The occurrence of TEM, SHV, and CTX-M together along with impermeability can cause resistance to carbapenems[25] which is a matter of concern in India which has high ESBL prevalence.
Conclusion
In our study, the 30.8% of GNB were ESBL producers. K. oxytoca was the most predominant ESBL organism followed by E. coli. Amikacin, colistin, tigecycline, and imipenem were the most effective antibiotics in ESBL-producing organisms in the present study. The rampant use of carbapenems for treating ESBL-producing organisms may lead to widespread carbapenem resistance. Bla TEM is the most predominant genotype in our locality in contrast to CTX-M which is prevalent in most parts of India. Ten percent cases in the present study had coexistence of all the genotypes which is a matter of concern because such isolates carry higher propensity for carbapenemase production as well.
Financial support and sponsorship
The study was financially supported by S'O'A University, Kalinga Nagar, Bhubaneswar, Odisha.
Conflicts of interest
There are no conflicts of interest.
References
- Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163-75.
- [Google Scholar]
- Molecular Cloning: A Laboratory Manual (2nd ed). Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989.
- Presence of genes for beta-lactamases of two different classes on a single plasmid from a clinical isolate of Serratia marcescens. J Antibiot (Tokyo). 1999;52:1135-9.
- [Google Scholar]
- Bla (CTX-M) genes in clinical Salmonella isolates recovered from humans in England and wales from 1992 to 2003. Antimicrob Agents Chemother. 2005;49:1319-22.
- [Google Scholar]
- Amp C beta-lactamase among ESBL-producing E.coli and If you don't look, you won't find. J Clin Diagn Res. 2009;3:1635-56.
- [Google Scholar]
- Variations in the prevalence of strains expressing an extended-spectrum beta-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin Infect Dis. 2001;32(Suppl 2):S94-103.
- [Google Scholar]
- Determining incidence of extended spectrum beta-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: The PEARLS study 2001-2002. Int J Antimicrob Agents. 2004;24:119-24.
- [Google Scholar]
- Genotypic detection of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a Tertiary care hospital. Int J Biomed Adv Res. 2015;6:91-7.
- [Google Scholar]
- Prevalence of extended-spectrum-beta-lactamase and metallo-beta-lactamase producing multi drug resistance gram – Negative bacteria from urinary isolates. Indian J Med Microbiol. 2013;31:420-1.
- [Google Scholar]
- CTX-M: Changing the face of ESBLs in the UK. J Antimicrob Chemother. 2005;56:451-4.
- [Google Scholar]
- Extended spectrum -lactamases in urinary isolates of Escherichia coli and Klebsiella pneumoniae – Prevalence and susceptibility pattern in a tertiary care hospital. Indian J Med Microbiol. 2004;22:172-4.
- [Google Scholar]
- Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS One. 2017;12:e0180121.
- [Google Scholar]
- Antibiotic resistance profile and extended spectrum beta-lactamase (ESBL) production in Acinetobacter species. Indian J Med Res. 2007;126:63-7.
- [Google Scholar]
- Plasmid-borne extended-spectrum beta-lactamase in a clinical isolate of Acinetobacter baumannii. J Med Microbiol. 2003;52:1125-7.
- [Google Scholar]
- High prevalence of PER-1 extended-spectrum beta-lactamase-producing Acinetobacter spp. In Korea. Antimicrob Agents Chemother. 2003;47:1749-51.
- [Google Scholar]
- Community acquired multi-drug resistant clinical isolates of Escherichia coli in a tertiary care centre of Nepal. Antimicrob Resist Infect Control. 2015;4:15.
- [Google Scholar]
- Frequency and antimicrobial sensitivity pattern of extended spectrum β-lactamases producing Escherichia coli and Klebsiella pneumoniae isolated in a tertiary care hospital. Al Ameen J Med Sci. 2010;3:265-71.
- [Google Scholar]
- Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol. 2011;29:161-4.
- [Google Scholar]
- Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in an urban hospital in Dhaka, Bangladesh. Int J Antimicrob Agents. 2004;24:508-10.
- [Google Scholar]
- Occurrence of the CTX-M, SHV and the TEM genes among the extended spectrum β-lactamase producing isolates of Enterobacteriaceae in a tertiary care hospital of North India. J Clin Diagn Res. 2013;7:642-5.
- [Google Scholar]
- Characterization of multidrug resistant extended-spectrum beta-lactamase-producing Escherichia coli among uropathogens of pediatrics in North of Iran. Biomed Res Int. 2015;2015:309478.
- [Google Scholar]
- Prevalence of TEM, SHV, and CTX-M beta-lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J Med. 2017;7:12-6.
- [Google Scholar]
- Bla (CTX-M), Bla (TEM), and Bla (SHV) in Enterobacteriaceae from North-Indian tertiary hospital: High occurrence of combination genes. Asian Pac J Trop Med. 2011;4:101-5.
- [Google Scholar]





