Translate this page into:
Performance Evaluation of an Improved HBsAg Assay (HBsAg NEXT) for the Detection of HBsAg Levels
Address for correspondence: Ekta Gupta, MBBS, MD (Microbiology), FRC Path (London), Department of Clinical Virology, Institute of Liver and Biliary Sciences, Vasant Kunj, New Delhi, 110070, India (e-mail: ektagaurisha@gmail.com; egupta@ilbs.in).
-
Received: ,
Accepted: ,
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objectives
Detection of hepatitis B surface antigen (HBsAg) plays an important role in the screening and diagnosis of hepatitis B virus (HBV) infections. There is a need of highly sensitive assays with an improved lower limit of detection (LoD). Here, we evaluate the performance characteristics of the HBsAg NEXT (HBsAg new) assay in the detection of HBsAg in clinical samples.
Materials and Methods
This was a cross-sectional study conducted at a tertiary care liver center in North India. The study included 439 clinical samples. The HBsAg new assay was compared to the conventional chemiluminescence-based assay (HBsAg old assay, Architect, Abbott Diagnostics, United States). The analytical sensitivity of the HBsAg new assay was evaluated by checking its performance with the second World Health Organization (WHO) international standards for HBsAg.
Results
Out of 439 blood samples that were retrieved from the departmental repository stored at –80°C, 100 samples were positive and 339 samples were negative for HBsAg as per the HBsAg old assay. The HBsAg new assay showed incremental detection of HBsAg in 11 additional samples. Out of these, 5 samples were confirmed as occult HBV infection and the remaining 6 were classified as “exposed-to-virus” samples (HBV core total antibody-positive). The HBsAg new assay demonstrated a high positive significant correlation with the HBsAg old assay (r = 0.881, p-value < 0.001). The HBsAg new assay could effectively detect the second WHO international standards to the level of 0.0033 IU/mL.
Conclusion
The HBsAg NEXT assay is a highly sensitive assay with an improved lower LoD.
Keywords
hepatitis B
immunoassay
HBsAg
occult hepatitis B
HBsAg NEXT
CHB
Introduction
Hepatitis B virus (HBV), a deoxyribonucleic acid (DNA) virus belonging to the Hepadnaviridae family, causes acute and chronic infections. Countries with a high HBV prevalence usually report incidences at an early age, which might lead to chronic infection. Countries with a low-to-intermediate prevalence report a high incidence in adulthood, which leads to acute and self-resolving illness, followed by viral clearance or, in rare cases, leading to acute liver failure.[1] Chronic hepatitis B (CHB) infection is a major public health problem.[1] Viral hepatitis is the seventh leading cause of death worldwide, with hepatitis B and C infections contributing to 96% of all hepatitis-related deaths.[2]
In India, the overall rate of hepatitis B surface antigen (HBsAg) positivity ranges from 1.1 to 12.2%, with approximately 40 million people chronically infected with HBV.[3] Approximately 25% of untreated patients of CHB develop complications such as cirrhosis or hepatocellular carcinoma.[3] The World Health Organization (WHO) Global Health Sector Strategy in 2016, declared elimination of viral hepatitis by 2030 as one of the major goals. The major bottlenecks in achieving the target of global control over viral hepatitis are extremely poor uptake of testing and lack of accurate and sensitive screening and diagnostic assays.[4]
Among different hepatitis B markers, HBsAg and HBV DNA are the most widely used markers for diagnosis and measurement of viral load in an active infection, respectively.[1,5] Over the years, several assays for detection of HBsAg have been developed with notable improvements in their sensitivity and specificity. However, several factors such as the presence of mutations in the HBV genome and low levels of antigen in the blood at different stages of infection limit their performance.[6,7,8] The diagnosis of occult HBV infection (OBI), which is defined as the absence of detectable HBsAg and the presence of HBV DNA in the blood, could be due to low levels of circulating HBsAg and low sensitivity of kits to detect the same.[9,10]
Therefore, new assays with higher sensitivity and lower limit of detection (LoD) are needed to overcome this challenge. Abbott ARCHITECT HBsAg NEXT (HBsAg new assay), a recently launched assay, is a one-step chemiluminescent microparticle immunoassay (CMIA) with an analytical sensitivity of 0.0052 IU/mL, that is, approximately equal to 0.02 ng/mL. This assay is performed on a fully automated Architect i2000 (Abbott, Wiesbaden, Germany) instrument.[11] There is limited literature available on its clinical utility. Therefore, in this study, we evaluated the performance characteristics of the HBsAg Next new qualitative assay in detecting HBsAg in clinical samples compared to that of the conventional ARCHITECT HBsAg Qualitative II assay (Abbott Diagnostics, Illinois, United States) (HBsAg old assay).
Materials and Methods
Study Design
This was a cross-sectional study conducted in the Department of Virology, Institute of liver and Biliary Sciences, New Delhi, India. The study was approved by the Institutional Ethics Board (IEC/2020/79/MA03) and conducted as per the principles of the Declaration of Helsinki. The study was performed on deidentified, anonymous, leftover archived clinical plasma samples. Hence, the requirement for individual patient consent forms was waived off.
Study Population
Plasma samples (within 3 months of testing), for which the results of HBsAg were available, were randomly retrieved from the departmental repository stored at –80°C. The samples were simultaneously tested by both the assays, that is, the HBsAg new and HBsAg old assays. All the samples were tested in the same freeze–thaw cycle. Blood samples that were coreactive for anti-human immunodeficiency virus (HIV) 1, HIV 2, and anti-hepatitis C virus (HCV) antibodies and those with uncertain storage conditions were excluded from the study. A total of 439 plasma samples were included in the study, of which 156 samples were from healthy voluntary blood donors and remaining were from patients. The retrieved samples were classified as per the disposition presented in ►Fig. 1.
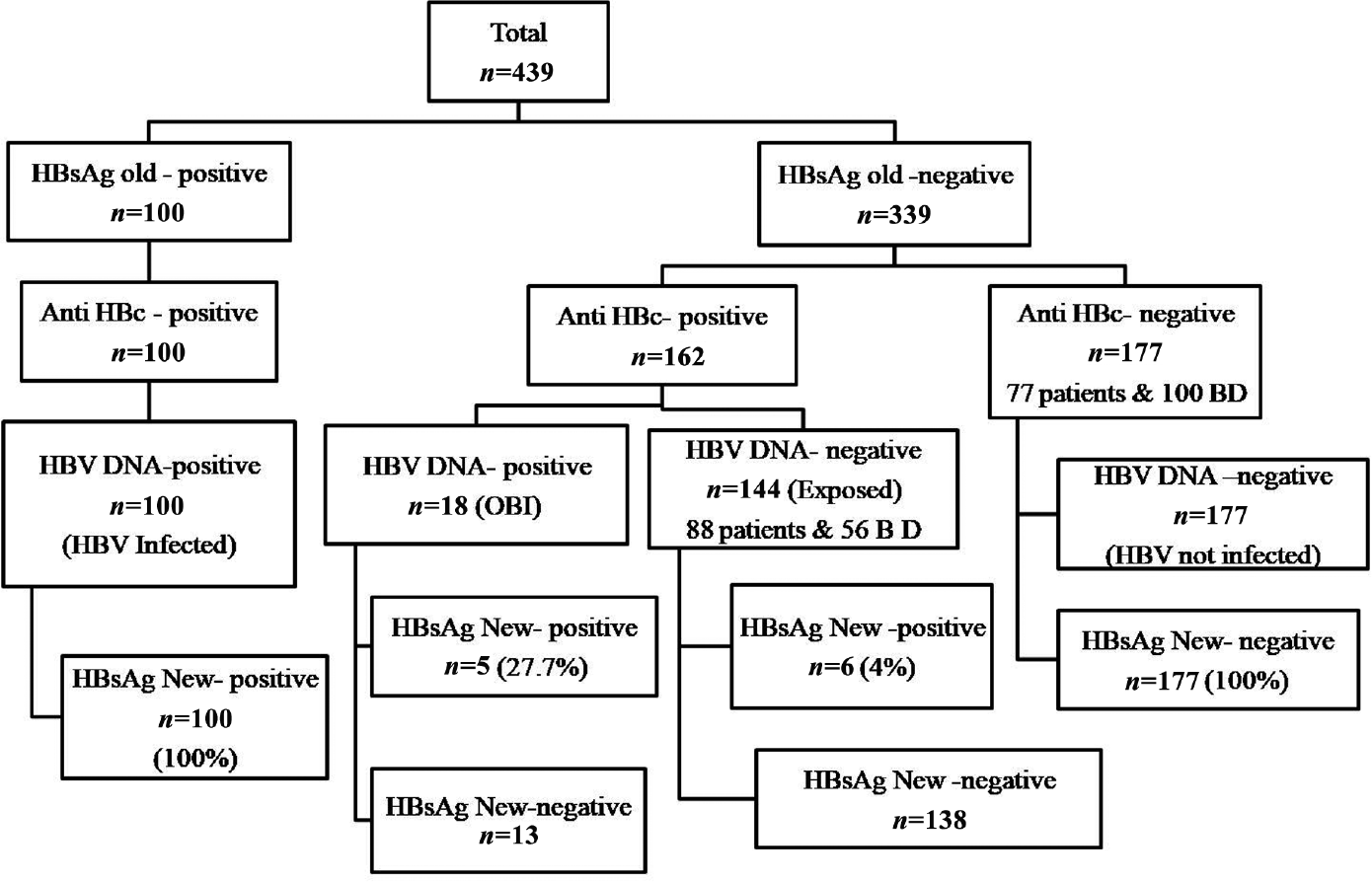
- The classification of the samples used in the study. BD, blood donors.
Study Definitions
HBV-infected: Samples positive for both HBsAg (old assay) and HBV DNA.
OBI: Samples negative for HBsAg (old assay) and positive for HBV DNA.
Exposed-to–infection: Samples negative for HBsAg (old assay) and negative for HBV DNA but positive for antibodies against the core antigen of HBV, that is, anti-hepatitis B core (HBc) total antibodies.
HBV-negative: Samples negative for all the three markers—HBsAg, HBV DNA, and anti-HBc total antibody.
Study Procedures
HBsAg Detection
HBsAg was detected in the clinical samples by both HBsAg new and HBsAg old assays. Both assays are commercially available as one-step chemiluminescent immunoassays and are used for the qualitative detection of HBsAg in human serum or plasma samples using CMIA technology as per the manufacturer's instructions. Sample, assay-specific diluents, anti-HBs coated paramagnetic microparticle, and anti-HBs acridinium-labeled conjugate are combined to create a reaction mixture and incubated. HBsAg, if present in the sample binds to anti-HBs microparticle and anti-HBs acridinium-labeled conjugate. After washing cycles and adding pre-trigger and trigger solutions, the resulting chemiluminescent reaction is read in relative light units. Both the assays were performed on the Abbott ARCHITECT i2000SR instrument.
Anti-HBc Total Antibody Detection
All the samples were tested for antibodies against the core antigen by ARCHITECT anti-HBc II assay as per the manufacturer's instructions. It is a chemiluminescent immunoassay for the qualitative detection of antibody to HBc antigen (anti-HBc) in human serum and plasma. It utilizes microparticle coated with recombinant HBV core antigen for the detection of anti-HBc. Anti-HBc determinations can be used as an indicator of current or past HBV infection.
HBsAg Confirmatory Assay
All discordant results of HBsAg were confirmed by HBsAg Next confirmatory assay for positivity as per the manufacturer's instructions. It is a chemiluminescent immunoassay for confirmation of the presence of HBsAg in human serum and plasma by means of specific antibody neutralization. It uses the principle of specific antibody neutralization to confirm the presence of HBsAg in samples found to be repeatedly reactive.
HBV DNA Estimation
All the samples were tested for HBV DNA using the COBAS AmpliPrep/COBAS TaqMan HBV test v2.0 assay on a COBAS AmpliPrep/COBAS TaqMan automated instrument as per the manufacturer's instructions, with a LoD as 10 IU/mL.
HBV Genotyping
HBV genotype information was available on a subset of samples (n = 27) with HBV DNA > 3 log10 IU/mL. HBV genotyping was done by Sanger sequencing of the surface gene.
(Forward primer: 5′-CATCAGGATTCCTAGGACCCCT-3′; Reverse primer: 5′-AGGACAAACGGGCAACATAC-3′).
Analytical Sensitivity of Assay
The analytical sensitivity of the HBsAg new assay was evaluated by checking its performance with the second WHO international standards for HBsAg, subtype adw2, genotype A containing 33 IU/mL. It was evaluated by running the standards in triplicate at different dilutions. The standards were first reconstituted and diluted in four sets of dilutions ranging from 101 to 104; these standards were then analyzed by the HBsAg new assay. All the dilutions were made in human plasma and they tested negative for the HBV, HCV, and HIV markers.
Statistical Analysis
Statistical analysis was performed using Statistical Package for the Social Sciences (SPSS) software version 2.2 (SPSS Inc., Chicago, United States). Observed statistical data were reported using standard techniques. Continuous variables such as age were summarized using mean with standard deviation (SD) or median and interquartile range, as applicable. Categorical variables were presented as percentages. A comparative evaluation of HBsAg old assay and HBsAg new assay was also performed. In addition, sensitivity, specificity, positive predictive value, and negative predictive value (NPV) were calculated for both assays—HBsAg new and HBsAg old assay. For statistical tests, a p-value of < 0.05 was considered significant.
Results
A total of 439 samples were included in the study, of which 100 samples were confirmed positive and the remaining samples were negative for HBsAg by the HBsAg old assay. Eighteen samples belonged to OBI. A total of 144 samples were classified as exposed-to-infection, out of which 88 samples were from patients and 56 samples were from asymptomatic blood donors. A total of 177 samples were true-negative samples, out of which 77 samples were from patients and 100 samples were from blood donors. The baseline characteristics are mentioned in ►Table 1.
| Category (n = 439) | HBsAg/HBc total/DNA positive (infected) (n = 100) | HBsAg/HBc total/DNA negative (not infected) (n = 77) | HBc total positive/DNA negative (exposed) (n = 88) | HBsAg negative/HBc total positive/DNA positive (OBI) (n = 18) | Blood donors (100 negative + 56 HBc total positive) (n = 156) |
|---|---|---|---|---|---|
| Age, mean (± SD), y | 42.4 ± 15.7 | 42.8 ± 14.8 | 49.4 ± 11.6 | 41.7 ± 10.3 | 38.6 ± 11.7 |
| M:F | 5:1 | 3:1 | 5:1 | 3:1 | 4:1 |
| Total bilirubin, mean (± SD), mg/dL | 2.8 ± 5.9 | 6.7 ± 9.7 | 1.4 ± 2.5 | 2.3 ± 3.4 | NA |
| Direct bilirubin, Mean (± SD), mg/dL | 1.3 ± 3.3 | 3.3 ± 5.3 | 1.1 ± 2.1 | 0.8 ± 1.2 | NA |
| ALT, mean (± SD), IU/L | 64.2 ± 146.2 | 209.2 ± 774.4 | 43.6 ± 68.18 | 34.2 ± 16.2 | NA |
| AST, mean (± SD), IU/L | 102.1 ± 283.8 | 196.0 ± 566.0 | 45.4 ± 182.4 | 22.1 ± 23.8 | NA |
Abbreviations: anti-HBc total, antibody to the core antigen of hepatitis B virus; ALT, alanine transaminase; AST, aspartate aminotransferase; DNA, deoxyribonucleic acid; F, female; HBsAg, hepatitis B surface antigen; M, male; OBI, occult HBV infection; SD, standard deviation.
Performance of the HBsAg New Assay
The HBsAg new assay accurately identified HBsAg in all the 100 confirmed positive samples and showed 100% concordance with confirmed HBsAg-negative samples, thereby demonstrating sensitivity and specificity of 100%. Interestingly, the HBsAg new assay showed incremental detection of HBsAg in 11 samples. All 11 samples were confirmed to be HBsAg-positive by the HBsAg Next confirmatory assay. Among the 11 samples, 5 samples belonged to OBI, in which both HBV DNA and anti-HBc total antibody were detected. In the remaining 6 of the 11 samples, only anti-HBc was detected. All 6 cases were that of chronic liver disease patients who were on antiviral treatment, which explained the low level of HBsAg and the HBV DNA-negative status (►Fig. 1).
Comparison of HBsAg New and HBsAg Old Assay
Both the assays showed excellent correlation with each other in the detection of HBsAg (r = 0.881, p-value < 0.001, ►Fig. 2). When sensitivity and specificity of both the assays were calculated based on HBV DNA and anti-HBc total results (as the gold standard assays for true positives) (►Table 2), the HBsAg new assay demonstrated a sensitivity of 89% compared to 85% for the HBsAg old assay. The NPV for the new assay was higher than that of the old assay.
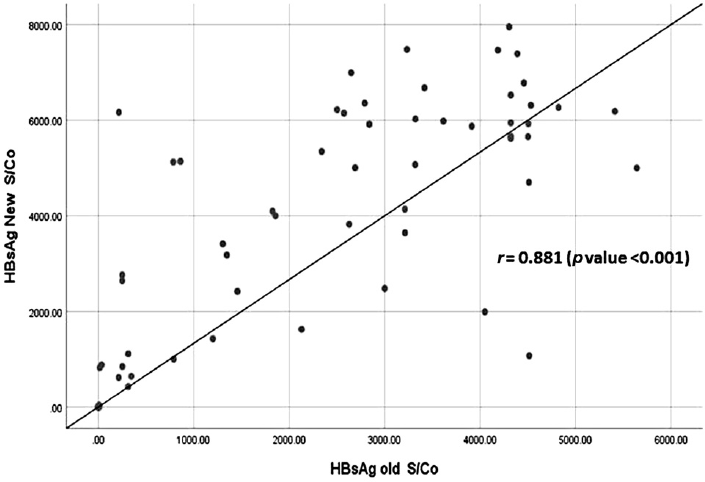
- A comparison between HBsAg new and HBsAg old assay in context to the detection of HBsAg.
| Category | HBsAg old (positive) | HBsAg new (positive) | Increased detection |
|---|---|---|---|
| Anti-HBc total and HBV DNA positive (true-positive), n = 118 | 100 | 105 | 5 |
| Anti-HBc total and HBV DNA negative (true-negative), n = 177 | 0 | 0 | 0 |
| Anti-HBc total positive and DNA negative (exposed), n = 144 | 0 | 6 | 6 |
| Sensitivity %, (95% CI) | 85 (77–91) | 89 (81–94) | – |
| Specificity %, (95% CI) | 100 | 100 | – |
| NPV %, (95% CI) | 91 (86.5–93.7) | 93.1 (89–95.8) | – |
| PPV, % | 100 | 100 | – |
| Accuracy %, (95% CI) | 92.6 (90.5–96.3) | 96 (92.5–97.6) | – |
Abbreviations: Anti-HBc total, antibody to the core antigen of HBV; CI, confidence interval; DNA, deoxyribonucleic acid; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; NPV, negative predictive value; PPV, positive predictive value.
Performance Evaluation HBsAg New and HBsAg Old Assay for OBI Detection
Out of 118 true-positive cases, the HBsAg old assay could detect HBsAg in 100 cases (84.7%), whereas the HBsAg new assay could detect it in 105 cases (88.9%) (►Table 2). Incremental increase was seen in OBI cases, that is, 5 out of 18 (27.7%). In 144 exposed patients; HBsAg new assay detected HBsAg additionally in 6 cases.
Analytical Sensitivity of the New Assay
The HBsAg new assay accurately detected the standard up to 1:10,000 dilutions, at a value of 0.0033 IU/mL. The HBsAg new assay demonstrated a good precision with all the values lying between ± 2 SDs (►Table 3).
| WHO serial dilution (IU/mL) | HBsAg new Result 1 (S/CO) | HBsAg new Result 2 (S/CO) | HBsAg new Result 3 (S/CO) | Range (± 2 SD) |
|---|---|---|---|---|
| WHO STD 1:1 | R (2,550.66) | R (2,311.42) | R (2,321.73) | 2,129.51–2,659.68 |
| WHO STD 1:10 | R (831.38) | R (554.01) | R (538.35) | 318.14–964.34 |
| WHO STD 1:100 | R (74.63) | R (75.87) | R (72.79) | 71.39–77.46 |
| WHO STD 1:1,000 | R (7.79) | R (7.75) | R (7.53) | 7.41–7.96 |
| WHO STD 1:10,000 | R (1.28) | R (1.22) | R (1.28) | 1.19–1.32 |
Abbreviations: HBsAg, hepatitis B surface antigen; R, reactive; S/CO: sample to cut-off; SD, standard deviation; WHO, World Health Organization.
Genotype-Wise Comparison
Genotyping data was retrieved for 27 samples. Genotype D was seen in 20 samples (74%), followed by genotype A in 6 samples (22.2%) and genotype C in 1 sample (3%). There was 100% concordance between the HBsAg new and HBsAg old assay in the detection of HBsAg among different genotypes.
Discussion
Monitoring serum/plasma HBsAg levels can substantially contribute to early detection among CHB individuals. Although multiple qualitative HBsAg detection assays are commercially available, a highly sensitive HBsAg detection assay is an absolute need of the hour. In our study, we evaluated the performances of the old ARCHITECT HBsAg Qualitative assay and newly launched ARCHITECT HBsAg Next assay in clinical samples. We demonstrated improved sensitivity of the HBsAg new assay in comparison to the HBsAg old assay. The results of the study indicated that the HBsAg new assay can detect HBsAg in OBI samples as well as treatment cases, wherein the HBV DNA is not detectable and antigen levels are very low. A similar kind of incremental detection and improved sensitivity was observed in previously published studies on this assay.[8,11]
HBsAg is the hallmark marker for screening as well as diagnosis of HBV infections[12] and newer assays with enhanced sensitivity and specificity are imperative. In terms of public health, assays with improved sensitivity aid in the diagnosis of HBV-infected patients who are in the low replication phase, earlier known as the inactive carrier stage. These patients are usually asymptomatic, and as the levels of HBsAg and HBV DNA are very low, detection by conventional assays can be challenging. Moreover, if these patients serve as blood donors, they can be an important vehicle for the transmission of transfusion-transmitted HBV infections. The WHO prequalification requirement for a screening assay for blood donation is that an assay should have a lower LoD < 0.130 IU/mL, with a sensitivity and specificity of 100 and > 98%, respectively. For a diagnostic assay, the requirement is that the test should have a LoD of < 4.0 IU/mL, with a sensitivity and specificity of 100 and > 98%, respectively.[13] In our study, the HBsAg Next assay, detected HBsAg antigen to a lower limit of 0.0033 IU/mL, thereby indicating enhanced sensitivity which would benefit blood screening safety.
In context to OBI, low levels of circulating HBsAg attributable to mutations in the “a” determinant region or pre-S1/pre-S2 regions of the virus, makes its detection challenging with the current diagnostic assays. Intriguingly, the HBsAg new assay could detect 5 out of 18 OBI samples (27.7%). It is very much possible that the remaining 13 OBI samples could have been missed by new HBsAg NEXT assay due to extremely low HBsAg levels below the detection limit of the assay or/and might have escape mutations not been able to be detected by the new assay. Globally, the prevalence of OBI varies from 1 to 87%.[14,15] Strict adherence to HBV DNA positivity is not maintained while defining OBI cases; the positivity for HBV DNA and anti-HBc total antibodies, which are markers of exposure to the virus, is often considered in parallel. In India, OBI is reported at a rate of up to 10.22% for anti-HBc positivity and 0.15% for HBV DNA positivity.[10,16] The screening of donated blood for HBV infections is not mandatory by nucleic acid testing (NAT) in most developing countries including India; thus, antigen assays with the capacity to detect low circulating levels of HBsAg are warranted to reduce the transmission of blood-borne HBV infections. Current immunoassays for the detection of HBsAg, such as enzyme-linked immunosorbent assay and the HBsAg old assay, have a LoD of 0.10 and 0.05 IU/mL, respectively. Currently, most diagnostic laboratories and blood banks are equipped with chemiluminescence-based assays for the detection of blood-borne viruses due to their high throughput, less turnaround time, and improved sensitivity and specificity. Therefore, adopting the new assay would not be a challenge for the diagnostic laboratories.
In conclusion, based on clinical performance evaluation, the HBsAg new assay was found to be satisfactory across genotypes and exhibited improved sensitivity and enhanced ability to detect OBI. This chemiluminescence-based assay would benefit diagnostic laboratories and blood banks for improved detection of HBV, especially in areas where infrastructure to set up NAT is not available.
There were certain limitations to our study. First that it was a retrospective cross-sectional study in which samples from the patients and healthy volunteers were examined at just one time point. Second, the study had a comparatively small sample size, multicenter studies with larger sample size are needed to strengthen our findings.
Consent for Publication
The study was performed on deidentified, anonymous, leftover archived clinical plasma samples. Hence, the requirement for individual patient consent forms was waived off.
Authors' Contributions
Conceptualization: E.G., M.K.S.; Methodology: E.G., A.K., A.B.; Data curation: A.R., A.B.; Validation: K.S., A.K.; Formal Analysis: A.R., A.B.; Funding acquisition: E.G.; Investigation: A.K., K.S., A.B.; Project administration: E.G; Resources: E.G.; Supervision: E.G.; Writing-original draft: E.G., A.B., A.K.; Writing, review, and editing: E.G., A.B., and J.S.
Acknowledgments
We would like to thank Abbott diagnostics for providing the mentioned kits for evaluation. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work, and have given their approval for this version to be published.
Conflict of Interest
None declared.
References
- Geneva: World Health Organization; 2017.
- National Guidelines for Diagnosis & Management of Viral Hepatitis. New Delhi, India: National Health Mission; 2018. p. :1-80.
- [Google Scholar]
- Lancet Gastroenterology & Hepatology Commissioners. Accelerating the elimination of viral hepatitis: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol Hepatol. 2019;4(02):135-184.
- [Google Scholar]
- Electronic address: easloffice@easloffice.eu, European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(02):370-398.
- [Google Scholar]
- Hepatitis B virus blood screening: need for reappraisal of blood safety measures? Front Med (Lausanne). 2018;5:29.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular and serological characterization of hepatitis B vaccine breakthrough infections in serial samples from two plasma donors. Virol J. 2019;16(01):43.
- [CrossRef] [PubMed] [Google Scholar]
- Performance characteristics of the high sensitivity Alinity i & ARCHITECT HBsAg Next Qualitative/Confirmatory assays. Diagn Microbiol Infect Dis. 2020;97(02):115033.
- [CrossRef] [PubMed] [Google Scholar]
- Hepatitis B viral safety of blood donations: new gaps identified. Ann Blood. 2018;3(38):1-5.
- [CrossRef] [Google Scholar]
- Update on occult hepatitis B virus infection. World J Gastroenterol. 2016;22(39):8720-8734.
- [CrossRef] [PubMed] [Google Scholar]
- An ultra-sensitive Abbott ARCHITECT® assay for the detection of hepatitis B virus surface antigen (HBsAg) J Clin Virol. 2018;105:18-25.
- [CrossRef] [PubMed] [Google Scholar]
- Improved detection of early acute, late acute, and occult hepatitis B infections by an increased sensitivity HBsAg assay. J Clin Virol. 2019;118(August):41-45.
- [CrossRef] [PubMed] [Google Scholar]
- Occult hepatitis B virus infection in a North American community-based population. J Hepatol. 2005;42(04):480-485.
- [CrossRef] [PubMed] [Google Scholar]
- High prevalence of occult hepatitis B virus genotype H infection among children with clinical hepatitis in west Mexico. Mem Inst Oswaldo Cruz. 2014;109(06):728-737.
- [CrossRef] [PubMed] [Google Scholar]
- Hepatitis B core antibody testing in Indian blood donors: a double-edged sword! Asian J Transfus Sci. 2012;6(01):10-13.
- [CrossRef] [PubMed] [Google Scholar]






