Translate this page into:
Genetic analysis of drug resistance mechanisms and phylogenetic clustering in Candida auris isolates from Western India
*Corresponding author: Shashikala Shivaprakash, Department of Microbiology, Sir HN Reliance Foundation Hospital and Research Centre, Mumbai, Maharashtra, India. shashikala.shivaprakash@rfhospital.org
-
Received: ,
Accepted: ,
How to cite this article: Chheda P, Shivaprakash S, Gupta N, Dama T, Biyani N, Bansode S. Genetic analysis of drug resistance mechanisms and phylogenetic clustering in Candida auris isolates from Western India. J Lab Physicians. 2024;16:447-53. doi: 10.25259/JLP_11_2024
Abstract
Objectives:
Candida auris is an emerging multidrug-resistant fungal pathogen that poses a significant threat to global health. Limited information is available from the Indian subcontinent regarding mutations associated with drug resistance and genetic variability among the isolates. In this study, we employed whole-genome sequencing (WGS) to investigate the genetic variations and drug resistance mechanisms within C. auris isolates from the western region of India.
Materials and Methods:
A total of twenty archived isolates were subjected to WGS on the Illumina NextSeq 2000 platform. A set of 18 genes was analyzed to check for the presence of drug-resistant mutations. Phylogenetic analysis was done using MEGA v6.06 software to identify the C. auris subgroup or clade and to check genetic relatedness among species.
Statistical analysis:
The data related to drug resistance were presented in numbers and percentages.
Results:
Through manual analysis, drug-resistant mutations were detected in ERG11, CDR1, and TAC1b genes, which are known to be associated with reduced susceptibility to antifungal agents. Phylogenetic analysis revealed that all the isolates clustered within Clade I, indicating a high degree of genetic similarity among isolates. The absence of comprehensive antifungal mutation databases and automated tools for drug resistance detection necessitated the utilization of specialized computational skills of bioinformaticians for data analysis.
Conclusions:
The study provides valuable insights into the genetic diversity and drug resistance mechanisms of C. auris isolates in the western region of India and emphasizes the need for continued research and surveillance to combat this emerging pathogen. Our findings underscore the need for the development of user-friendly automated tools and comprehensive databases to facilitate rapid and accurate identification of drug resistance in C. auris.
Keywords
Bioinformatics
Candida auris
Whole genome sequencing
INTRODUCTION
Candida auris is an emerging fungal pathogen that has recently gained attention due to its ability to cause severe and often fatal infections in hospitalized patients. First identified in 2009 in Japan, C. auris has since been reported in over 40 countries, and its global spread has raised concerns among healthcare professionals and public health authorities. C. auris is a multidrug-resistant organism that is particularly adept at surviving on hospital surfaces and equipment, making it difficult to control and eradicate.[1] In addition, C. auris infections can be difficult to diagnose, as they often do not respond to standard antifungal treatments and can be mistaken for other types of infections.[2]
C. auris has an average genome size of approximately 12.5 megabases (Mb) and is known to have multiple clades or distinct genetic lineages. As of now, five major clades have been identified based on genomic analysis - Clades I (South Asian), II (East Asian), III (South African), IV (South American), and V (Iranian) are genetically distinct. Clades I– IV were simultaneously and independently identified in South Asia, East Asia, Africa, and South America, respectively. [3,4]
One of the key mechanisms of antifungal resistance in C. auris is the overexpression of drug efflux pumps, which reduces the concentration of the drug inside the fungal cell and renders it less effective. Similarly, mutations in the ERG11 gene (lanosterol 14-alpha-demethylase enzyme) have been associated with reduced susceptibility to azoles, another class of antifungal medications commonly used to treat Candida infections.[5] Another mechanism of antifungal resistance in C. auris is the presence of mutations in genes involved in the synthesis or modification of the fungal cell wall. For example, mutations in the FKS1 gene have been associated with reduced susceptibility to echinocandins, a class of antifungal medications that inhibit the synthesis of beta-glucan, a key component of the fungal cell wall.[6] Other genes associated with antifungal resistance in C. auris include the CDR1 and MDR1 genes, which encode for efflux pumps that can transport a broad range of drugs out of the fungal cell, and the TAC1 gene, which encodes for a transcription factor that regulates the expression of multiple efflux pump genes.[6,7]
Mutations in these genes can lead to increased expression of efflux pumps and decreased susceptibility to antifungal medications. In addition to these genetic mechanisms of antifungal resistance, C. auris has also been associated with biofilm formation, which can protect the fungus from host defenses and antifungal medications.
C. auris has caused outbreaks in healthcare facilities in India. The first case of C. auris was reported in India in 2011, and since then, it has become a significant public health concern due to its high mortality rate and difficulty in treating infections.[8] C. auris infections have been reported in several states in India, and the prevalence of the pathogen is believed to be underreported due to inadequate laboratory testing and surveillance.[9]
Rapid and accurate identification of drug-resistant strains and the determination of clade relationships are critical for effective outbreak control and patient management. This was an observational study aimed to utilize whole-genome sequencing (WGS) to investigate drug resistance and clade identification in C. auris isolates. In addition, phylogenetic analysis was done to determine clade relationships among the isolates, which would provide valuable insights into the epidemiology and evolution of this emerging pathogen.
MATERIALS AND METHODS
A total of 20 archived isolates (8 clinical and 12 screening for colonization at axilla and groin sites) were sequenced for whole genome analysis. The 8 clinical isolates comprised of urine (n = 5), ascetic fluid (n = 1), bronchoalveolar lavage (n = 1), and endotracheal fluid (n = 1). Of the 12 screening samples undertaken for the study, 8 were groin swabs, and 4 were axilla swabs. All the isolates were previously identified by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry (VITEK MS platform) as well as by Sanger sequencing of the internal transcribed spacer (ITS) region. The study was conducted at Sir HN Reliance Foundation Hospital, Mumbai.
Deoxyribonucleic acid (DNA) extraction
The genomic DNA was isolated from 200 µL of colony suspension using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Species’ identities were confirmed by sequencing the ITS region. The extracted DNA was quantified using Qubit dsDNA BR Assay (Invitrogen, Carlsbad, USA) on Invitrogen™ Qubit™ 2.0 Fluorometer as per the manufacturer’s instructions. The DNA quality (OD 260/280) was determined on a Nanodrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Library preparation and next generation sequencing
Library preparation was carried out using the Illumina DNA Prep Kit as per the manufacturer’s instructions. Sequencing was performed using NextSeq 1000/2000 P1 Reagents (300 Cycles) on the NextSeq 2000 platform (Illumina, San Diego, CA, USA), creating 2 × 150 bp paired-end reads.
Data analysis
FASTQ files were imported to FastQC v0.11.7 for assessment of read quality. Since the reads were of good quality, trimming was not required. The reads were then used for de novo assembly using SPAdes v3.6.0 assembler, and 20 supercontigs each were generated for 20 isolates. A variant calling file (VCF) was generated using B11221 as the reference genome. Variants were identified using Genome Analysis Tool Kit (HaplotypeCaller, v3.7-0) and RealignerTargetCreator, IndelRealigner, HaplotypeCaller for both single nucleotide polymorphisms (SNPs) and insertions and deletions (indels). Sites were filtered with Variant Filtration using “QD < 2.0 | FS > 60.0 | MQ < 40.0”. The VCF file was annotated with snpEff v5.0e and filtered using SnpSift v5.0e.[10,11]
Gene prediction
Based on the final assemblies and VCF files of the isolates, proteins and coding genes were predicted using the AUGUSTUS v3.2.1 tool on the GALAXY platform. Once the GFF files were obtained, putative ORFs were searched manually against the NR database of NCBI (http://www.ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) and Candida genome database (http://www.candidagenome.org/).
Antifungal mutation analysis
Data obtained from GFF files were further used for detecting resistance mutations in a set of 18 genes associated with drug resistance by aligning gene sequences of individual isolates against the B11221 reference genome using ClustalW alignment (MEGA v6.06 software). The data related to drug resistance was presented in numbers and percentages.
Phylogenetic analysis
For the identification of clades and to visualize genetic relatedness among isolates, phylogenetic analysis was carried out using 20 assembled sequences (fasta sequences) and 5 reference genomes representing different clades (Clade I - B8441, Clade II - B11220, Clade III - B11221, Clade IV - B11245, and Clade V - IFRC2087). Genome-wide SNP-based phylogenetic tree was constructed using the maximum parsimony using bootstrap analysis with 1000 reiterations algorithm in MEGA V6.06 software.[12]
Antifungal susceptibility testing
Antifungal susceptibility testing was conducted on eight clinical isolates of C. auris as per the Centers for Disease Control and Prevention criteria using the Sensititre YeastOne YO10 colorimetric antifungal panel. The isolates were cultured on Sabouraud dextrose agar and incubated at 35°C for 24–48 h. A standardized inoculum was prepared by suspending colonies in sterile saline to achieve a turbidity equivalent to a 0.5 McFarland standard. The YeastOne panels were inoculated with the standardized suspension and incubated at 35°C for 24 h. Minimum inhibitory concentrations (MICs) were determined based on the colorimetric change in the wells, following the manufacturer’s guidelines.
RESULTS
In our study, we employed WGS to investigate the genetic variations within C. auris. The analysis of next-generation sequencing (NGS) data from C. auris requires specialized bioinformatic skills and access to appropriate computational resources to unravel the genetic characteristics and potential drug-resistant mechanisms.
The NextSeq 2000 run on the P1 flow cell yielded a total of 46.40 gigabase (Gb) data with 161.44 million reads (92.9% at ≥ Q30). The total number of reads passing the filter was 139.10 million, and an average of 2.3 Gb of data (average sequencing depth > × 160) was generated per sample. Each of the 20 isolates had a genome size of approximately 12.2 Mb. The raw data generated out of whole genome sequencing for all 20 isolates have been deposited in the NCBI Sequence Read Archive under the accession ID PRJNA1049194. (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1049194)
Antifungal-resistant mutations
Given the absence of comprehensive antifungal mutation databases specific to C. auris, several known antifungal resistance-associated genes were manually analyzed including CDR1 CDR2, CIS2, ERG2, ERG3, ERG6, ERG11, ERG13, FCY1, FKS1, FKS2, FUR1, MDR1, MEC3, MRR1A, PEA2, TAC1a, and TAC1b. The analysis involved a cluster-wise alignment approach, as no automated tools for fungi currently exist.
Among the analyzed isolates, 18 (90%) exhibited the Y132F mutation in the ERG11 gene, a known marker of azole resistance. In addition, these 18 isolates showed the E709D mutation in the CDR1 gene, which has been associated with multidrug resistance [Table 1]. The remaining two (S02 and S04) isolates displayed distinct resistance profiles, harboring the K143R mutation in the ERG11 gene, the V704L mutation in the CDR1 gene, and the A640V mutation in the TAC1b gene.
| Isolate No. | Source | ERG11 | CDR1 | TAC1b | Predicted drug resistance |
|---|---|---|---|---|---|
| S01 | Axilla | Y132F | E709D | - | Fluconazole |
| S02 | Axilla | K143R | V704L | A640V | Fluconazole |
| S03 | Urine | Y132F | E709D | – | Fluconazole |
| S04 | Urine | K143R | V704L | A640V | Fluconazole |
| S05 | Groin | Y132F | E709D | – | Fluconazole |
| S06 | Groin | Y132F | E709D | – | Fluconazole |
| S07 | Urine | Y132F | E709D | – | Fluconazole |
| S08 | Groin | Y132F | E709D | – | Fluconazole |
| S09 | Urine | Y132F | E709D | – | Fluconazole |
| S10 | Groin | Y132F | E709D | – | Fluconazole |
| S11 | Ascitic fluid | Y132F | E709D | – | Fluconazole |
| S12 | Urine | Y132F | E709D | – | Fluconazole |
| S13 | Endo tracheal fluid | Y132F | E709D | – | Fluconazole |
| S14 | Axilla | Y132F | E709D | – | Fluconazole |
| S15 | Axilla | Y132F | E709D | – | Fluconazole |
| S16 | Bronchoalveolar lavage | Y132F | E709D | – | Fluconazole |
| S17 | Groin | Y132F | E709D | – | Fluconazole |
| S18 | Groin | Y132F | E709D | – | Fluconazole |
| S19 | Groin | Y132F | E709D | – | Fluconazole |
| S20 | Urine | Y132F | E709D | – | Fluconazole |
In addition to the known drug-resistant mutations, the presence of several SNPs and small insertions/deletions (indels) in various genes across all 20 C auris isolates was revealed in this study [Figure 1]. However, the functional significance of these genetic variations in C. auris remains unknown.
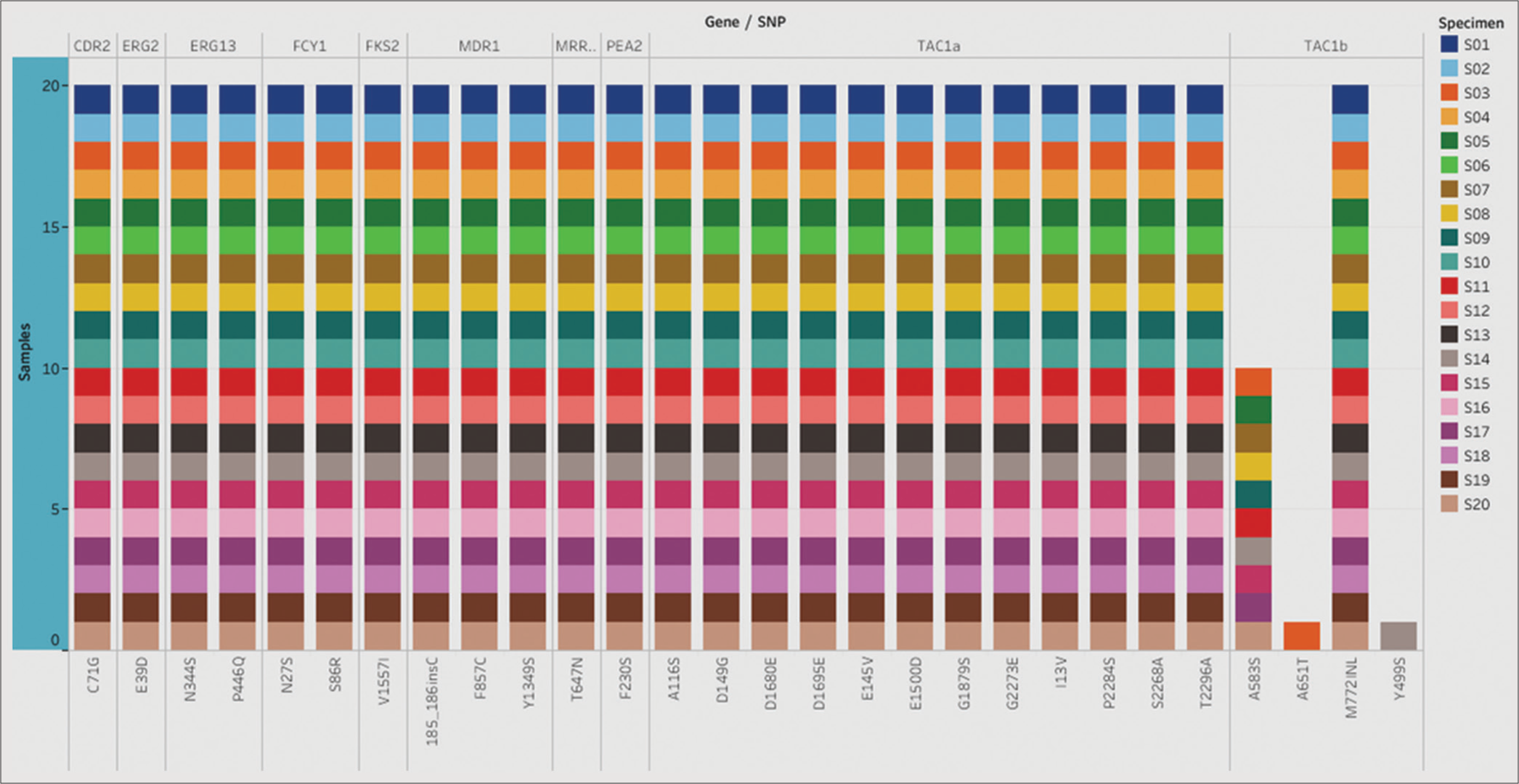
- List of single nucleotide polymorphism (SNPs) identified in the selected genes. The stacked plot illustrates SNPs identified in corresponding genes in all 20 Candida auris isolates.
Antifungal susceptibility
Antifungal susceptibility testing was performed on the isolates (n = 8) derived from clinical specimens [Table 2]. All eight clinical isolates exhibited resistance to fluconazole, while three isolates demonstrated resistance to amphotericin B and three to caspofungin.
| Isolates | FLZ (µg/mL) | AMP B(µg/mL) | CAS(µg/mL) | MCF (µg/mL) | AND (µg/mL) | Mutations |
|---|---|---|---|---|---|---|
| S03 Urine | R (>256) | S | S | S | S | ERG11 Y132F, CDR1 E709D |
| S08 Urine | R (>256) | S | S | S | S | ERG11 Y132F, CDR1 E709D |
| S09 Urine | R (32) | R (8) | S | S | S | ERG11 Y132F, CDR1 E709D |
| S11 Ascitic fluid | R (>256) | R (4) | R (>8) | S | S | ERG11 Y132F, CDR1 E709D |
| S12 Urine | R (32) | S | S | S | S | ERG11 Y132F, CDR1 E709D |
| S13 Endo tracheal fluid | R (32) | R (8) | S | S | S | ERG11 Y132F, CDR1 E709D |
| S16 BAL fluid | R (>256) | S | R (>8) | S | S | ERG11 Y132F, CDR1 E709D |
| S20 Urine | R (>256) | S | R (>8) | S | S | ERG11 Y132F, CDR1 E709D |
R: Resistant, S: Susceptible, FLZ-Fluconazole, AMP B: Amphotericin B, CAS: Caspofungin, MCF: Micafungin, AND: Anidulafungin
Phylogenetic study
In this study, we conducted a comprehensive phylogenetic analysis utilizing the powerful MEGA software to examine the clade as well as genetic relatedness among 20 C. auris isolates. These isolates were compared to five reference clades to determine their evolutionary relationships. Our results revealed that all the isolates exhibited a striking resemblance to clade I, indicating a high degree of similarity in their genetic profiles [Figure 2]. These isolates of C. auris were further divided into subclades: 1b (18/20) and 1c (02/20), characterized by the presence of ERG11 Y132F and ERG11 K143R mutations, respectively.
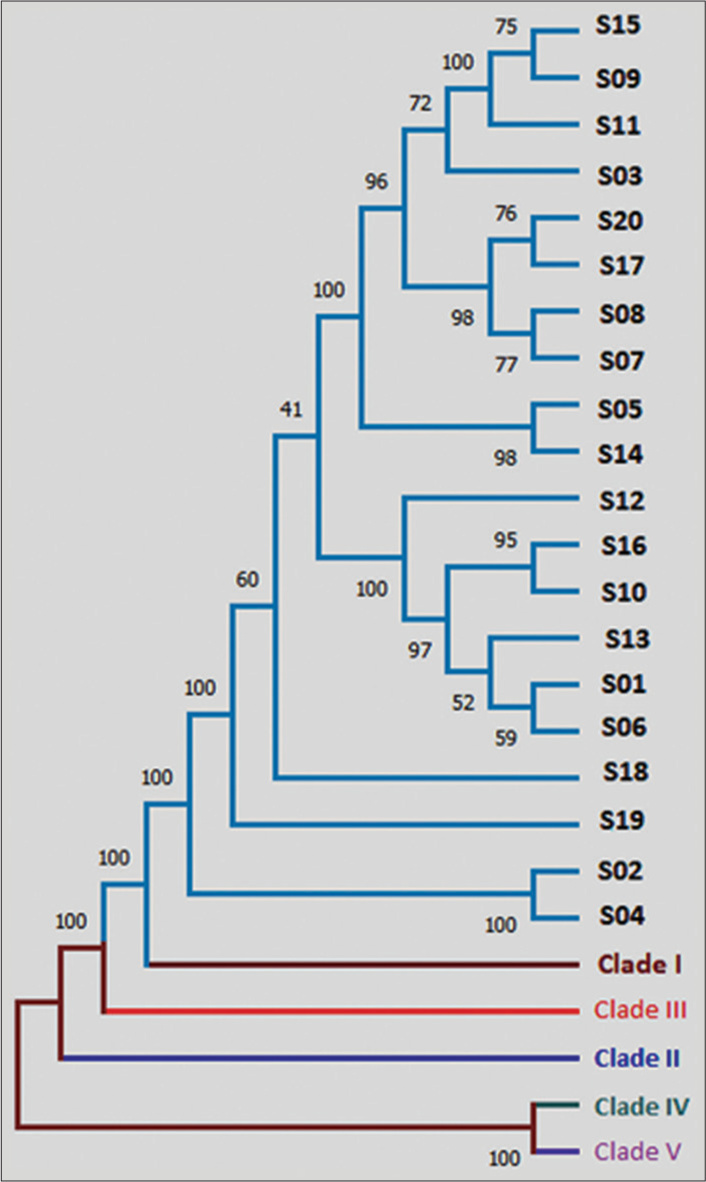
- Phylogenetic analysis using MEGA v6.06 software. Phylogenetic analysis revealed close resemblance of all 20 isolates to Clade I.
Another aspect was to understand the relatedness among these isolates based on the output derived from phylogenetic analysis. There were observable similarities between some isolates, e.g., S05 and S14, S02 and S04 [Figure 2]. The transmission dynamics of C. auris could not be conclusively determined in this study due to the random selection of isolates. However, the use of phylogenetic analysis provides a valuable tool to assess the degree of relatedness among different C. auris species.
DISCUSSION
NGS data analysis for C. auris typically involves several steps, such as quality control, read alignment, variant calling, and annotation. These steps aim to identify genetic variations, including SNPs, insertions, deletions, and structural variants, within the C. auris genome. The analysis of 20 C. auris isolates also included a comparison of the obtained genomic data with reference sequences or databases to understand the genetic diversity and potential mechanisms of drug resistance in these strains.
The identification of the Y132F mutation in the ERG11 gene is a well-documented genetic alteration associated with reduced susceptibility to azole antifungal drugs (fluconazole), which are commonly used in the treatment of Candida infections. It has been previously shown that Y132F leads to a ≥4-fold rise in fluconazole MIC. [13] Two of our strains showed the presence of K143R mutation, which significantly increases fluconazole and voriconazole MIC by 8–16 fold.[14] Another important ERG11 mutation, VF125AL, which imparts fluconazole resistance, was not identified in our study. VF125AL is uniquely identified in a subclade under the South African clade (Clade III).[14,15] The presence of either Y132F or K143R mutations in all our isolates suggests a potential challenge in managing C. auris infections, as it may limit the effectiveness of azole therapy. Antifungal susceptibility testing was performed on eight of the clinical C. auris isolates, all of which showed resistance to fluconazole with MIC of >32 mg/L, and it correlated with the presence of ERG11 Y132F mutation in all.
CDR1 gene mutations were detected across all 20 isolates in our study. Both the mutations, E709D and V704I, associated with azole resistance, have been found in C. auris strains recovered from apples in an Indian study.[16] The CDR1 protein plays a crucial role in the efflux of antifungal agents from the cell, contributing to reduced drug accumulation and therapeutic failure.[17] The presence of this mutation highlights the need for alternative treatment strategies and underscores the importance of ongoing surveillance to monitor the emergence and spread of multidrug-resistant C. auris strains.
Another finding was the detection of A640V mutation in the TAC1b gene in two (10%) of the isolates. TAC1b is known to regulate the expression of efflux pumps, which play a key role in mediating azole drug resistance in C. auris.[14] It has been demonstrated that TAC1b promotes CDR1 expression, and deletion of TAC1b results in decreased resistance to azoles.[7]
The two isolates (S02 and S04) in our study displaying the K143R mutation in the ERG11 gene, the V704L mutation in the CDR1 gene, and the A640V mutation in the TAC1b gene represent unique resistance profiles and highlight complex mechanisms of resistance in C. auris.
Many studies have highlighted that CDR1 and TAC1b play an important role in the efflux of antifungal drugs; however, it is not explicitly mentioned to which set of drugs it imparts resistance. Besides known drug-resistant mutations, several SNPs were identified; though the majority of these could be clade-specific variations, the association of some of these variants with drug resistance cannot be ruled out.[14,16,17]
The susceptibility testing results of eight clinical samples are summarized in Table 2. Notably, all samples exhibited resistance to fluconazole, corroborating previous reports of widespread resistance in C. auris to this antifungal agent. In addition, isolates S09 and S13 demonstrated resistance to amphotericin B, isolates S16 and S20 to caspofungin, and isolates S11 to both amphotericin B and caspofungin. These results underscore the significant challenge posed by multidrug resistance in this pathogen. The observed correlation between antifungal resistance and the presence of genotypic resistance markers, such as ERG11, CDR1, and TAC1b, suggests a potential association between specific genetic mutations and resistance development in C. auris. These findings highlight the urgent need for continued surveillance and the development of alternative therapeutic strategies to manage infections caused by this emerging pathogen.
The absence of FKS1 mutations in our study of 20 C. auris isolates from the western region of India highlights a notable finding. FKS1 mutations are typically associated with echinocandin resistance in C. auris. However, the absence of these mutations suggests either susceptibility to echinocandins among these isolates or the existence of alternative mechanisms contributing to echinocandin resistance specific to this population in the western region of India. Previous multicentric studies conducted in different regions of India have reported FKS1 mutations, indicating potential regional variations in the genetic profile of C. auris isolates and their associated antifungal resistance mechanisms.[18,19]
Fluconazole resistance associated with mutations in ERG11, CDR1, and TAC1b emphasizes the urgent need for continued surveillance and the development of alternative therapeutic strategies to manage infections caused by this emerging pathogen effectively.
The identification of clade I or South Asian strain in our Indian hospital isolates is consistent with the other studies from the Indian subcontinent, and this information adds to the growing body of evidence on the global distribution and genetic diversity of C. auris.[20,21] The identification of clade I as the predominant clade suggests a potential regional or local transmission pattern within the studied population. While the random selection of isolates in this study limits the direct assessment of transmission dynamics, the application of phylogenetic analysis enhances our understanding of the relatedness among C. auris species. By examining the evolutionary relationships inferred from this analysis, researchers can indirectly infer potential transmission events and inform future investigations and control measures to combat the spread of this emerging pathogen.
CONCLUSIONS
Our study of 20 C. auris isolates, using WGS, revealed that all isolates belonged to clade I. Drug-resistant mutations were detected in genes such as ERG11, CDR1, and TAC1b. The absence of an antifungal mutation database or automated tools for drug resistance detection highlights the reliance on the computational skills of bioinformaticians for analysis.
Moving forward, the development of comprehensive antifungal mutation databases and user-friendly automated tools for drug resistance detection would greatly enhance the efficiency and accessibility of such analyses. This would increase our understanding of the genetic mechanisms driving antifungal resistance in C. auris and facilitate rapid identification and monitoring of drug resistance in C. auris isolates, aiding in the implementation of effective treatment strategies and infection control measures.
Ethical approval
This is an observational study performed on archived yeast isolates and patient consent or ethics committee approval has not been taken. The study strictly adheres to institutional guidelines for retrospective analyses, ensuring compliance with all relevant regulations and ethical standards. The authors’ take complete responsibility for the same.
Declaration of patient consent
Patient’s consent is not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Candida auris A pathogen difficult to identify, treat, and eradicate and its characteristics in Japanese strains. J Infect Chemother. 2019;10:743-9.
- [CrossRef] [Google Scholar]
- Identification of drug resistant Candida auris. Front Microbiol. 2019;10:1918.
- [CrossRef] [Google Scholar]
- Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25:1780-1.
- [CrossRef] [Google Scholar]
- Clade distribution of Candida auris in South Africa using whole genome sequencing of clinical and environmental isolates. Emerg Microbes Infect. 2021;10:1300-8.
- [CrossRef] [Google Scholar]
- Drug resistance and novel therapeutic approaches in invasive Candidiasis. Front Cell Infect Microbiol. 2021;11:759408.
- [CrossRef] [Google Scholar]
- Combined antifungal resistance and biofilm tolerance: The global threat of Candida auris. mSphere. 2019;4:e00458-19.
- [CrossRef] [Google Scholar]
- A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere. 2020;5:e00279-20.
- [CrossRef] [Google Scholar]
- Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 2015;41:285-95.
- [CrossRef] [Google Scholar]
- A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80-92.
- [CrossRef] [Google Scholar]
- Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35.
- [CrossRef] [Google Scholar]
- MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725-9.
- [CrossRef] [Google Scholar]
- Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother. 2015;59:450-60.
- [CrossRef] [Google Scholar]
- Delineation of the direct contribution of Candida auris ERG11 mutations to clinical triazole resistance. Microbiol Spectr. 2021;9:e0158521.
- [CrossRef] [Google Scholar]
- Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: A three-year surveillance study. Emerg Microbes Infect. 2021;10:1088-96.
- [CrossRef] [Google Scholar]
- Candida auris on apples: Diversity and clinical significance. mBio. 2022;13:e0051822.
- [CrossRef] [Google Scholar]
- Functional expression of recombinant Candida auris proteins in Saccharomyces cerevisiae enables azole susceptibility evaluation and drug discovery. J Fungi (Basel). 2023;9:168.
- [CrossRef] [Google Scholar]
- A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73:891-9.
- [CrossRef] [Google Scholar]
- Impact of FKS1 genotype on echinocandin in vitro susceptibility in Candida auris and in vivo response in a murine model of infection. Antimicrob Agents Chemother. 2022;66:e0165221.
- [CrossRef] [Google Scholar]
- Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio. 2021;12:e03181-20.
- [CrossRef] [Google Scholar]
- Colonisation and transmission dynamics of Candida auris among chronic respiratory diseases patients hospitalised in a chest hospital, Delhi, India: A comparative analysis of whole genome sequencing and microsatellite typing. J Fungi. 2021;7:81.
- [CrossRef] [Google Scholar]






