Translate this page into:
Alarming emergence, molecular characterization, and outcome of blaNDM-1 in patients infected with multidrug-resistant Gram-negative bacilli in a tertiary care hospital
Address for correspondence: Dr. Meher Rizvi, Department of Microbiology, JNMC, AMU, Aligarh, Uttar Pradesh, India. E-mail: rizvimeher@gmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
INTRODUCTION:
This study was conducted to assess the prevalence of metallo-beta-lactamases (MBLs) in general and blaNDM-1 in particular. It also aimed at evaluating clinical characteristics and outcome in patients infected with MBLs.
MATERIALS AND METHODS:
A total of 116 carbapenem-resistant Gram-negative bacilli (CRGNB) were evaluated in the study. These CRGNB were tested for MBL production both phenotypically for MBLs and genotypically for blaNDM-1 gene by polymerase chain reaction (PCR). Representative stains of NDM-1 isolates were further sequenced by Triyat Scientific Co., (Nagpur, India).
RESULTS:
Among 116 CRGNB Citrobacter species 28 (24.13%) was the most common pathogen. Phenotypically, MHT, imipenem-EDTA (IPM-EDTA) double-disk synergy test and IPM-EDTA combined disk synergy test (CDST) detected MBL production in 105 (90.51%), 96 (81.03%), and 87 (75%) CRGNB, respectively. However, blaNDM-1 genes were detected in 66 (56.89%) isolates. The prevalence of blaNDM-1 gene was highest among Escherichia coli 26 (100%). Considering PCR as gold standard, it was observed that IMP-EDTA CDST was most specific (78.38%) while MHT was most sensitive (97.47%). Results of blaNDM-1 gene by PCR were further confirmed by sequencing (Triyat genomics, Nagpur). All the 11 representative strains were confirmed to be an NDM-1 gene. The presence of MBLs in our group of patients (non-Intensive Care Unit patients) is a cause for concern. However, on tracing their outcome, it was interesting to note that while the duration of stay lengthened in a large number of patients 112 (96.5%), mortality was relatively low 5 (4.31%).
CONCLUSION:
The results of this study provide insight into the prevalence of MBLs, including blaNDM-1, in a tertiary care hospital. Antibiotic stewardship implemented in all seriousness may to a great extent stave off the impending pan-drug resistance. The surprising outcome of our patients suggests either that the bacteria trade off virulence for drug resistance or the relatively robust immune response of non ICU patients fights back.
Keywords
blaNDM-1
carbapenem-resistant Gram-negative bacilli
metallo-beta-lactamase
sequencing
Introduction
The global dissemination of acquired metallo-beta-lactamases (MBLs) in Gram-negative bacteria is a clarion call for the emergence of the postantibiotic era.[1] Most of the transposable MBLs encoding genes are carried as cassettes on integrons which facilitate their rapid spread among organisms and confer resistance to both beta-lactams and other antimicrobial agents. MBL encoding genes have been detected from several Gram-negative bacilli (GNB) belonging to the family Enterobacteriaceae and also in Pseudomonas species and Acinetobacter species.[2] Until now, multiple allelic variants of several MBLs have been described (http://www.lahey.org/studies/); blaVIMs and blaIMPs are the most frequent MBLs worldwide[3] and outbreaks of blaVIM and blaIMP type MBLs in Pseudomonas aeruginosa have been reported from Greece, Italy, Korea, and China.[4567] Recently, a novel metalloenzyme, New Delhi MBL-1 (NDM-1), was discovered in Klebsiella pneumoniae in a Swedish patient subsequent to treatment in a hospital in New Delhi, India[8] and has since then disseminated rapidly globally.[9101112] NDM-1 shares only 32.4% amino acid sequence homology with the closely related VIM-1/VIM-2 MBL producers and with the exception of other MBLs, transposes readily to other bacteria by ISCR1 element via rolling circle replication.[8] Because this carbapenemase is encoded by a genetic element found on different plasmids that may duplicate or jump from bacteria to bacteria easily, rapid dissemination, and spread between different bacterial species by lateral gene transfer favored by globalization and travel, represent a high risk of a worldwide pandemia among Gram-negative pathogens.
Considering the grave public health threat the MBL producers pose, we undertook this study to assess the prevalence of MBLs in general and blaNDM-1 in particular. We also aimed to evaluate clinical characteristics and outcome in patients infected with MBLs.
Materials and Methods
The present study was conducted in patients admitted to wards or attending the OPD of J. N. Medical College and Hospital, AMU, Aligarh in the Department of Microbiology from February 2014 to December 2015. This study was done after approval from Institutional Ethics Committee of J. N. Medical College and the procedures followed in the study were in accordance with its guidelines. Fluid samples were obtained from intercostal chest tube drainage (ICTD), peritoneal fluids, and pleural fluids. Clinical specimens including pus, urine, and fluid submitted to the bacteriology were investigated. The samples were obtained with proper aseptic techniques and transported to the laboratory within 1 h of their collection. The organisms were identified on the basis of cultural characteristics, morphology and biochemical tests (Bailey and Scotts, 2007,[13] Mackie and McCartney, 2007[14]).
Antibiotic susceptibility testing was performed by Kirby Bauer disc diffusion technique on Mueller-Hinton agar according to CLSI 2010, M 100-S20[15] guidelines. However, certain other drugs apart from CLSI 2010 guidelines were also tested. Antimicrobial disks used were as follows:
For Enterobacteriaceae (pus and fluids)
First-line drugs
Amikacin (30 μg), gentamicin (10 μg), levofloxacin (5 μg), ofloxacin (5 μg), ceftriaxone (30 μ g), cefoperazone (75 μg), cefotaxime (30 μg), cefixime (5 μg), cefepime (30 μg), ceftazidime (30 μg), cefpodoxime (10 μg), cefoperazone-sulbactam (75/10 μg), ceftazidime-clavulanic acid (30/10 μg), ceftriaxone-sulbactam (30/15 μg), and cefotaxime-clavulanic acid (30/10 μg).
Second-line drugs
Ceftazidime-tazobactam (80/10 μg), piperacillin (100 μg), piperacillin + tazobactam (100/10 μg), fosfomycin (200 μg) tobramycin (10 μg), sparfloxacin (5 μg), ertapenem (10 μg), faropenem (5 μg), and imipenem (10 μg).
For nonfermenting Gram-negative bacilli
First-line drugs
Amikacin (30 μg), gentamicin (10 μg), levofloxacin (5 μg), ceftriaxone (30 μg), cefoperazone (75 μg), cefepime (30 μg), nitrofurantoin (300 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefpodoxime (10 μg), cefoperazone-sulbactam (75/10 μg), and ceftriaxone-sulbactam (30/15 μg).
Higher drugs
Ceftazidime-clavulanic acid (30/10 μg), ticarcillin (75 μg), piperacillin (100 μg), piperacillin + tazobactam (100/10 μg), tobramycin (10 μg), sparfloxacin (5 μg), imipenem (10 μg), colistin (10 μg), polymyxin B (300 units).
For urinary tract infection the drugs used were as follows
First-line drugs
Amikacin (30 μg), gentamicin (10 μg), levofloxacin (5 μg), ceftriaxone (30 μ g), cefoperazone (75 μg), cefoperazone-sulbactam (75/10 μg), cefixime (5 μg), cefepime (30 μg), nitrofurantoin (300 μg), ceftazidime (30 μg).
Second line drugs
Ceftazidime-tazobactam (80/10 μg), piperacillin (100 μg), piperacillin + tazobactam (100/10 μg), tobramycin (10 μg), sparfloxacin (5 μg), ertapenem (10 μg), faropenem (5 μg), and imipenem (10 μg).
Consecutive, nonduplicate, carbapenem-resistant GNB (CRGNB) isolates were screened phenotypically for detection of MBL production. Confirmation of MBL production was done by polymerase chain reaction (PCR). Detailed history and investigations of all these patients were noted and followed up for outcome.
Phenotypic detection of metallo-beta-lactamase production
Modified Hodge Test, imipenem-EDTA (IPM-EDTA) double-disk synergy test (DDST) and IPM-EDTA combined disk synergy test (CDST) were utilized for phenotypic detection of MBL
Modified Hodge Test for metallo-beta-lactamase detection
Modified Hodge Test was performed as described by Lee et al., 2001. An inoculum of Escherichia coli ATCC 25922, equivalent to the 0.5 McFarland turbidity standard, was used to prepare a lawn culture on Mueller-Hinton agar plate, by swabbing with a sterile cotton swab. The test strain was heavily streaked from center to periphery. After the plate was allowed to stand for 15 min at room temperature, a 10 μg imipenem disk (Hi media, India) was placed at the center, and the plate was incubated overnight. The presence of a distorted inhibition zone (“cloverleaf shaped”) was interpreted as a positive result for carbapenem hydrolysis, and the strains were phenotypically screened for MBL production.
Imipenem-EDTA double-disk synergy test
IPM-EDTA DDST was performed as described by Lee et al., 2001.[16] Inoculum with turbidity equivalent to 0.5 McFarland standard was prepared. MHA plates were inoculated by swabbing them with a sterile cotton swab. With a sterile forceps, 10 μg imipenem disk (Hi media, India) and a plain disk was placed at a distance of 10 mm (edge to edge) on the MHA plate. 10 μl of 0.5 M EDTA solution was dropped over the plain disk. Plates were incubated at 35°C for 18–24 h. After overnight incubation, the presence of a synergistic inhibition zone was interpreted as a positive result for MBL production.
Imipenem-EDTA combined disk synergy test
IPM-EDTA CDST was performed as described by Yong et al., 2009.[8] Inoculum with turbidity equivalent to 0.5 McFarland standard was prepared. MHA plates were inoculated by swabbing them with a sterile cotton swab. With a sterile forceps, two 10 μg imipenem disks (Hi media, India) were placed on the plate, and 10 μL of 0.5 M EDTA solution was inoculated on one of them. Plates were incubated at 35°C for 18–24 h. The inhibition zones of the imipenem and IPM-EDTA disks were compared after 16-18 h of incubation in air at 35°C. In the combined disc test, if the increase in inhibition zone with the imipenem and EDTA disc was ≥5 mm than the imipenem disc alone, it was considered as MBL positive.
Genotypic detection of blaNDM-1
Genotypic detection of blaNDM-1 genes was performed only on those isolates which were phenotypically identified as MBL producers. Thus, one hundred sixteen isolates were selected for detailed molecular characterization.
DNA extraction for blaNDM-1
Bacterial DNA template was prepared from freshly cultured bacterial strains by scraping 2–3 colonies from the culture plate and emulsifying/suspending them in 400 μl of Tris-EDTA buffer pH 8.0 and then heating at 95°C for 10 min, followed by immediately chilling at 4°C. This was followed by centrifugation at 13,000 rpm for 5 min. About 300 μl of supernatant was pipetted out and stored at − 20°C till further use.
Methodology for amplification of blaNDM-1
All study isolates were subjected to PCR. PCR amplification was carried out on a Gradient Thermo Cycler named Le cycler of LABNIC, USA using primers targeting blaNDM-1.
Primers design
Primers were ordered from Eurofins Genomics Pvt., Ltd., Bengaluru India.
NDM-1 Primers - The primers used for the amplification of the NDM-type of MBLs at a conc of 10 picomolar each which amplified a 621 bpamplicon.[17]
-
NDM-Fm 5’-GGTTTGGCGATCTGGTTTTC-3’
-
NDM-Rm 5’-CGGAATGGCTCATCACGATC-3’.
The master mix used was bought commercially from FERMENTAS, Life sciences, USA. Standardization of reaction mixture for blaNDM-1 was done using a different amount of master mix, primers, and template several times. Cycling condition was also standardized by applying varying gradients of annealing temperature and different numbers of cycles in the amplification of MBL genes, to get the best results. The final reaction mixture and cycling conditions are mentioned here.
The PCR program carried out in thermal cycler with an initial denaturation step at 94°C for 10 min followed by 34 cycles of DNA denaturation at 94°C for 40 s, primer annealing at 52°C for 40 s and primer extension at 72°C for 50 s with a holding temperature of 72°C for 7 min. After amplification, end product was run on Agrose gel by electrophoresis. 1% agarose (Bangalore Genei, India) gel containing ethidium bromide was used and observed under the gel doc system from Bio-rad (Australia).
Sequencing
Representative PCR products of NDM-1 isolates were purified and sequenced by Triyat Scientific Co., (Nagpur, India), using forward and reverse primers as were used for PCR. Sequencing reactions were performed in an MJ Research PTC-225 Peltier Thermal Cycler using a ABI PRISM® BigDyeTM Terminator Cycle Sequencing Kits with AmpliTaq® DNA polymerase (FS enzyme) (Applied Biosystems), following the protocols supplied by the manufacturer. Single-pass sequencing was performed on each template using Universal primer. The fluorescent-labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI 3730xl sequencer (Applied Biosystems).
Phylogenetic analysis
For sequence alignment as well as phylogenetic analysis, we selected the GenBank sequences with the best and the high-scoring matches with NDM-1 reported sequences. Sequences were edited, aligned and analyzed using Clustal W Bio-edit software. Genetic distances were calculated using the maximum composite likelihood algorithm and phylogenetic trees was constructed by the neighbor-joining method. To confirm the reliability of the pairwise comparison and phylogenetic tree analysis, bootstrap sampling, and reconstruction were carried out 1000 times. Phylogenetic analysis was done using MEGA version MEGA 4 package.
Urinary tract infection (UTI) was defined as positive quantitative urine culture (>105 microorganisms/ml) with a maximum of two isolated microbial species in patients with symptoms suggestive of UTI. Wound infection was defined as per Southampton grading. Majority of the cases belonged to Grade III and IV. Fluid cultures were interpreted according to Bergey's Manual of Systematic Bacteriology Volume 3.
All CRGNB positive patients were kept isolated from the other patients and all the hospital staff were advised to practice transmission-based precautions and other universal precautions with more conscious attention to hand hygiene
Results
A total of 116 consecutive, nonduplicate CRGNB were included in the study, out of which 85 (73.27%) were isolated from pus, 17 (14.65%) from urine and 14 (12.06%) from fluid. Citrobacter species 28 (24.13%) predominated followed by E. coli 26 (22.41%), Pseudomonas species 24 (20.68%), Serratia species 17 (14.65%) Klebsiella species 9 (7.75%), Acinetobacter species 5 (4.31%), Proteus species 4 (3.44%), Providencia species 2 (1.72), and Aeromonas species 1 (0.86%). These CRGNB were further tested for MBL production both phenotypically and genotypically.
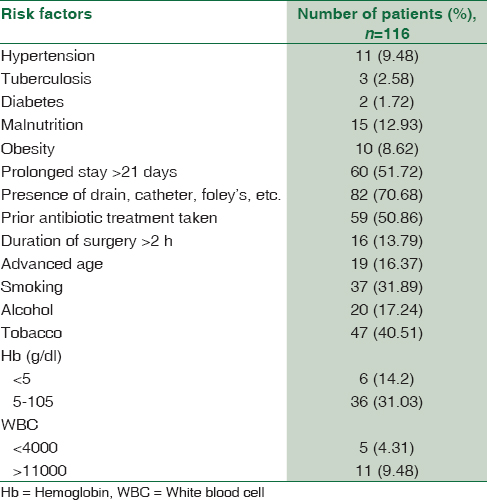
Table 1 describes the risk factors associated with hospitalized patients. It was observed that 82 (70.68%) patients had in dwelling devices such as an intravenous catheter and urinary catheters, 60 (51.72%) patients had prolonged stay in hospital >21 days, and 59 (50.86%) patients were on prior antibiotic treatment. Risk factors associated with advanced age, surgery for >2 h duration and malnutrition were lower 18 (15.51%), 15 (12.93%), and 16 (13.79%) respectively. It was further noticed that obesity, hypertension, diabetes, and tuberculosis were least associated risk factors with hospitalized patients, in this study.
Modified Hodge Test
It was observed that 105 (90.51%) carbapenem resistant gram negative bacilli (CRGNB) isolates were MBL positive by Modified Hodge Test. Predominant MBL producers were Citrobacter species 25 (23.80%), E. coli 25 (23.80%) and P. aeruginosa 23 (21.90%) followed by Serratia species 15 (14.28%) and Klebsiella species 8 (7.61%). Unequivocal result was obtained in 9.48% isolates, among which predominant were Citrobacter species 27.27% and Serratia species 18.18%.
Double-disk synergy test
As compared to Modified Hodge Test, fewer CRGNB isolates 96 (81.03%) were identified as MBL positive by IMP-EDTA DDST. Predominant MBL producer was again Citrobacter species 22 (23.40%), E. coli 22 (23.40%) and P. aeruginosa 20 (21.27%) positive for MBL results followed by Serratia species 15 (15.95%) and Klebsiella species 9 (6.38%). Unequivocal result was obtained in 22 (18.96%) isolates, among which predominant were Citrobacter species 27.27%.
Combined disk synergy test
Only 87 (75.00%) CRGNB strains were identified as MBL positive by IMP-EDTA CDST. Predominant MBL producer was Citrobacter species 23 (26.43%) followed by E. coli 19 (21.83%) and P. aeruginosa 17 (19.54%), followed by Serratia species 13 (14.94%) and Klebsiella species 6 (6.89%). Unequivocal result was obtained in 29 (25.00%) Gram-negative isolates.
Thus phenotypically Citrobacter species was the predominant MBL producer, followed by E. coli and Pseudomonas species. Surprisingly, very few of MBL producers were observed in Klebsiella Species.
Polymerase chain reaction for blaNDM-1
Of the 116 isolates, blaNDM-1 genes were detected in 66 (56.89%) isolates. They were obtained from pus 46 (69.69%), urine 10 (15.15%) and fluid (15.15%). Although the most common pathogen isolated in the study was Citrobacter species 28 (24.13%), the prevalence of blaNDM-1 gene was highest among E. coli 26 (100%) isolates [Figure 1].
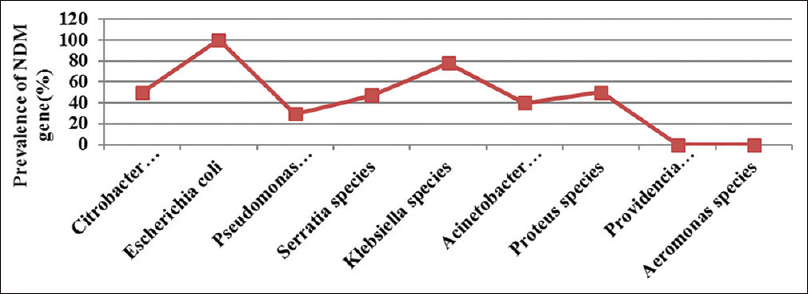
- Prevalence of blaNDM-1 gene among bacterial pathogens
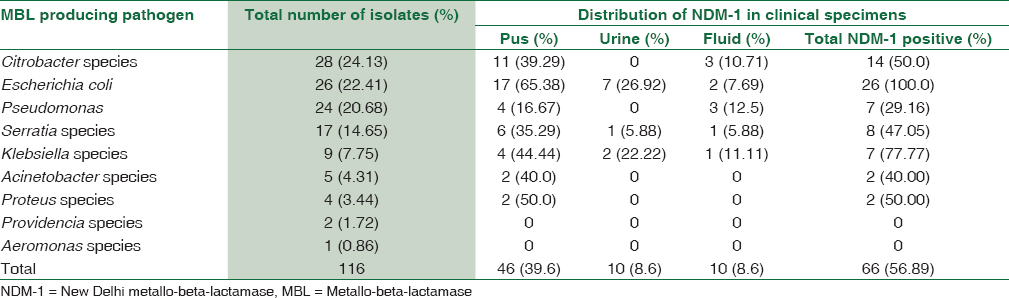
The most common blaNDM-1 bacterial isolate in pus were E. coli 17 (65.38%) and Citrobacter species 11 (39.29%). The predominant blaNDM-1 bacterial isolates in urine were again E. coli 7 (26.92%) and Klebsiella species 2 (22.22%). However, in fluids P. aeruginosa and Citrobacter species 3 (30.0%) each were predominant blaNDM-1 isolates [Table 2]. Surprisingly, all the E. coli 26 (100%) isolates were blaNDM-1 producing isolates whereas none of the isolates of Providencia and Proteus species were blaNDM-1 positive.
On analyzing the phenotypic and genotypic results, it was observed that IMP-EDTA CDST most closely correlated with the genotypic results. Sensitivity and specificity of Modified Hodge Test, IMP-EDTA DST, IMP-EDTA CDST was calculated with PCR as gold standard. It was observed that IMP-EDTA CDST was most specific (78.38%) while MHT was most sensitive (97.47%).
Direct sequencing of blaNDM-1 gene
Results of blaNDM-1 gene by PCR were further confirmed by sequencing (Triyat genomics, Nagpur). All the 11 representative strains (DNA extraction No. 90, 97, 103, 110, 111, 115, 116, 119, 139, 159, 176) were confirmed to be NDM-1 gene. Figure 2 displays the phylogenetic tree. Figure 3 shows 2% agarose gel electrophoresis results of PCR for the detection of blaNDM-1 gene.
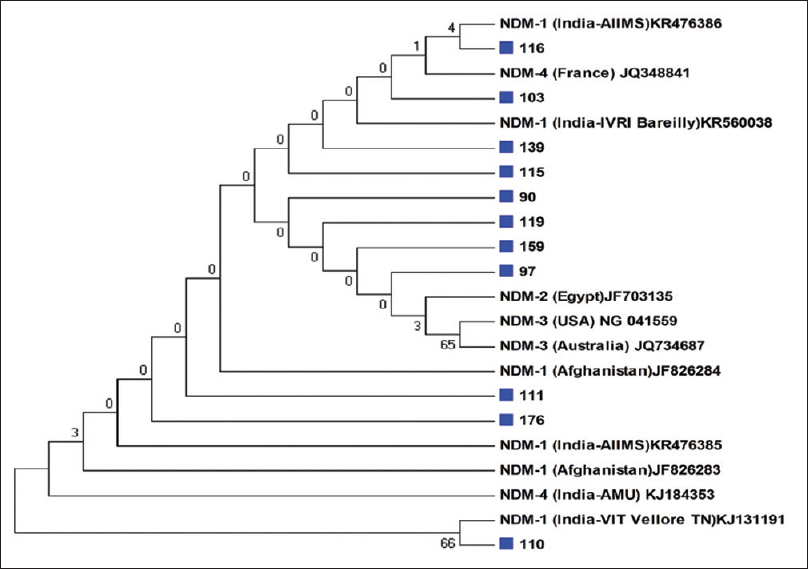
- Study samples belonging to New Delhi metallo-beta-lactamase-1 strains are marked with blue color squares in our study population
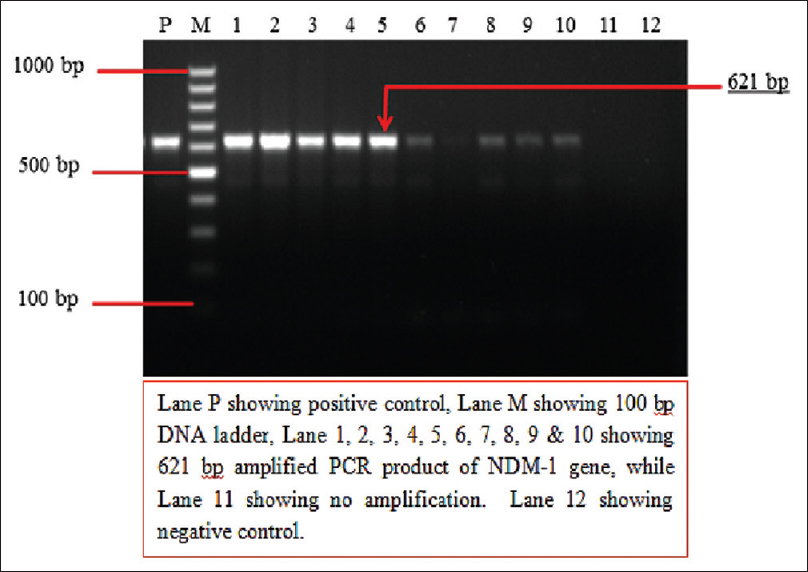
- Two percent agarose gel electrophoresis showing results of polymerase chain reaction for the detection of blaNDM-1gene
Discussion
The prevalence of multidrug resistance (MDR) among GNB has increased alarmingly. Compared with infections due to antimicrobial-susceptible GNB, infections due to MDR-GNB lead to worse outcomes, longer hospital stays, increased mortality, and greater costs of hospitalization.[18]
Antimicrobial-resistant pathogens historically have been traditionally considered to be primarily nosocomial. However, it is now evident that these pathogens have spread not only to other health care settings but have also made substantial inroads into the community resulting in an influx of patients who have antimicrobial-resistant pathogens isolated at hospital admission itself.[19]
In our center, Citrobacter species 28 (24.13%) was the predominant CRGNB. It was surprising that prevalence of carbapenem resistant Klebsiella species 9 (7.75%) was considerably lower than E. coli 26 (22.41%).
In this study, it was observed that 90.51% isolates were identified as MBLs by Modified Hodge test while IPM-EDTA DDST detected 81.03% isolates were MBL positive and IMP-EDTA CDST identified 75% MBLs. Predominant MBL producers by all three methods were Citrobacter species followed by E. coli and P. aeruginosa.
On comparing the prevalence of NDM-1by genotypic methods (56.89%) and the three phenotypic tools, it was observed that detection by IMP-EDTA CDST was the most discriminatory and specific (75.00%). Thus, among the standard phenotypic detection tests, the most specific was CDST (75%), and MHT was the most sensitive (90.51%). EDTA-based tests appear to be more specific. We recommend IMP-EDTA CDST for confirmation of MBLs.
Debasrita et al., 2011[20] compared phenotypic and genotypic methods for detection of MBLs. They observed that 26% bacteria showed false positives test by phenotypic method. They also suggested that for screening MBL producing bacteria, three phenotypic methods should be used before giving positive results, and one should not rely solely only on one method (especially DDST). However, for confirmation three methods should be followed by PCR.
While phenotypically, Citrobacter spp. was the predominant MBL producer, genotypically, it was conclusively established that E. coli was the predominant MBL producer in general (unpublished data) and blaNDM-1 in particular. It was surprising that while K. pneumoniae was phenotypically a low MBL producer, genotypically it was the second largest blaNDM-1producer.
Overall, predominant blaNDM-1 genes were observed in E. coli (100%) followed by Klebsiella species (77.77%) and Citrobacter species (50%). Surprisingly, all MBL producers in urine sample were blaNDM-1. The most common blaNDM-1 producing bacterial isolates in pus were Citrobacter species 78.57%, Serratia species 75%, E. coli 65.38 followed by P. aeruginosa and Klebsiella species (57.14%) each. In UTIs, on the other hand, the predominant bacterial isolates were E. coli 26.92%, Klebsiella species 22.22%, followed by Serratia species 5.88%. Again in fluids, the etiology varied with P. aeruginosa 12.5% predominating followed by Klebsiella species 11.11% and Citrobacter species 10.71%. Similar findings were observed by Gheorghe et al., 2014.[21]
The high prevalence of CRE phenotype of Citrobacter and Serratia spp. may be due to hyperproduction of AmpC. This suggests that hyperproduction of AmpC also gives a positive MHT. We suggest that in case of Citrobacter and Serratia spp., a positive phenotypic test need not suggest MBL production. They should be further subjected to genotypic detection.
blaNDM-1 has emerged as the major MBL gene with a high prevalence of 56.89% in Aligarh region of Western U.P. This enzyme was first identified in Sweden from a tourist returning from India. Since then, there has been a spate of reports from many countries, indicating its rapid and alarming dissemination. Amudhan et al., 2011[22] too reported that blaNDM-1 was the most prevalent MBL (57.65%). In other Indian studies, the prevalence of blaNDM-1 producers among carbapenem-resistant Enterobacteriaceae ranged between 31.2% and 91.6%.[2223] The high prevalence of blaNDM-1 gene in our study was confirmed by sequencing (Triyat genomics, Nagpur). All the 11 representative strains were confirmed to be NDM-1 gene.
NDM-1 is located on plasmids of different sizes and location. They also have high transmissibility and disseminate among other species leading to their spread in E. coli, K. pneumoniae, Enterobacter species, Citrobacter freundii, Morganella species, and Providencia species. Several studies have indicated extensive genotypic heterogeneity in the NDM-1 producing E. coli and K. pneumoniae.[24] In our study too, blaNDM-1 was prevalent in all species barring Providencia and Aeromonas species.
The majority of patients recovered (P < 0.001). This suggests that MBLs/NDM-1 may have low virulence or they may even be innocent bystanders. However, it must be kept in mind that this was a study in patients admitted to wards (not seriously ill patients), thus their immune response may play a significant role in recovery. However, recovery did take time as is manifested by the increased duration of hospital stay (P < 0.001). Thus, in a non-Intensive Care Unit patient, morbidity due to MBL infection increased, however, mortality was significantly low. Kus et al., 2011, reported that NDM-1 though a resistant pathogen can remain as a colonizer for 3–5 months without producing clinical infection despite the presence of multiple comorbidities in the patient.[25] Chen et al., 2011[26] also arrived at a similar conclusion.
Notwithstanding the fact that genotypic tests are gold standards for detecting MBLs, it is also true that in resource-constrained settings, molecular tests are not available, are expensive, technologically demanding and labor intensive. Results of our study suggest that EDTA CDST most closely mirrors the genotypic prevalence of MBL in general and NDM-1 in particular and should be preferred over MHT and EDTA DDST. We recommend that EDTA CDST should be utilized routinely as it is easy to perform, discriminatory, reproducible and cheap tool for detection of MBLs.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 2007;74:1686-701.
- [Google Scholar]
- Metallo-beta-lactamases: The quiet before the storm? Clin Microbiol Rev. 2005;18:306-25.
- [Google Scholar]
- Beta-lactamases identified in clinical isolates of Pseudomonas aeruginosa. Crit Rev Microbiol. 2010;36:245-58.
- [Google Scholar]
- Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000;38:1290-2.
- [Google Scholar]
- Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-beta-lactamase. Clin Infect Dis. 2000;31:1119-25.
- [Google Scholar]
- Outbreak by meropenem-resistant Pseudomonas aeruginosa producing IMP-6 metallo-beta-lactamase in a Korean hospital. Diagn Microbiol Infect Dis. 2009;63:115-7.
- [Google Scholar]
- Outbreak of Pseudomonas aeruginosa producing IMP-9-type metallo-beta-lactamase in Guangzhou, China. Int J Antimicrob Agents. 2008;32:363-5.
- [Google Scholar]
- Characterization of a new metallo-beta-lactamase gene, bla (NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046-54.
- [Google Scholar]
- Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597-602.
- [Google Scholar]
- New Delhi metallo-ß-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg Infect Dis. 2011;17:103-6.
- [Google Scholar]
- Emergence of metallo-ß-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother. 2010;54:4914-6.
- [Google Scholar]
- Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother. 2011;55:934-6.
- [Google Scholar]
- Bailey and Scott's Diagnostic Microbiology (12th ed). Mosby, Elsevier; 2007. 11830 Westline Industrial Drive, St. Louis, Missouri 63146
- Mackie & McCartney. Practical Medical Microbiology (14th ed). Churchill Livingstone: Publshed by Elsevier, a division of Reed Elsevier India Private Limited; 2014.
- Clinical Laboratory Standards Institute. Performance Standard for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement M200-S20. Wayne, PA, USA: Clinical Laboratory Standards Institute; 2010.
- Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7:88-91.
- [Google Scholar]
- Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791-8.
- [Google Scholar]
- Rapidly rising prevalence of nosocomial multidrug-resistant, gram-negative bacilli: A 9-year surveillance study. Infect Control Hosp Epidemiol. 2004;25:842-6.
- [Google Scholar]
- Preventing the influx of vancomycin-resistant enterococci into health care institutions, by use of a simple validated prediction rule. Clin Infect Dis. 2004;39:964-70.
- [Google Scholar]
- Study on some Gram-negative multi drug resistant bacteria and their molecular characterization research article. Asian J Pharm Clin Res. 2011;4:108-12.
- [Google Scholar]
- Molecular screening of carbapenemase-producing gram-negative strains in Romanian intensive care units during a one year survey. J Med Microbiol. 2014;63:1303-10.
- [Google Scholar]
- Bla(IMP) and bla(VIM) mediated carbapenem resistance in Pseudomonas and Acinetobacter species in India. J Infect Dev Ctries. 2012;6:757-62.
- [Google Scholar]
- Detection and characterization of metallo beta lactamases producing Pseudomonas aeruginosa. Indian J Med. 2015;28:241-4.
- [Google Scholar]
- Increasing prevalence and dissemination of NDM-1 metallo-ß-lactamase in India: Data from the SMART study (2009) J Antimicrob Chemother. 2011;66:1992-7.
- [Google Scholar]
- New Delhi metallo-ß-lactamase-1: Local acquisition in Ontario, Canada, and challenges in detection. CMAJ. 2011;183:1257-61.
- [Google Scholar]
- Spontaneous eradication of a NDM-1 positive Klebsiella pneumoniae that colonized the intestine of an asymptomatic carrier. J Chin Med Assoc. 2011;74:104.
- [Google Scholar]





