Translate this page into:
Association of intercellular adhesion gene A with biofilm formation in staphylococci isolates from patients with conjunctivitis
Address for correspondence: Prof. Mona Saad Nour, 5 Adeeb Sebaay Street, Nozha Gadeeda, Cairo, Egypt. E-mail: monasaadnour@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
There is a great negative impact of biofilm-mediated infection on patient health which necessitates the use of reliable methods for detecting biofilm producers.
AIMS:
This study was done to determine biofilm-producing ability and the presence of intercellular adhesion gene A in clinical staphylococcal isolates and to assess the reliability of two phenotypic methods used for biofilm detection.
MATERIALS AND METHODS:
Fifty staphylococcal strains were isolated from 100 conjunctival swabs from patients attended the Ophthalmology Outpatient Department of the Research Institute of Ophthalmology. Two phenotypic methods were used for detection of biofilm production; qualitative congo red agar (CRA); and quantitative microtiter plate. Polymerase chain reaction was used to determine the presence of icaA gene.
RESULTS:
In Staph aureus, 60% were positive biofilm forming and 40% were negative biofilm forming by both phenotypic methods. All positive biofilm-forming isolates were positive for icaA gene production. In coagulase negative staph, 50% were positive biofilm forming and 50% were negative biofilm forming by both phenotypic methods. All positive biofilm-forming strains were positive for icaA gene. All negative cases by CRA and microtiter plate methods were negative for icaA gene except two isolates. All staphylococcal isolates were subjected to antibiotic susceptibility test to correlate biofilm formation with multidrug resistance in staph.
CONCLUSION:
There is high significant correlation between icaA gene presence and biofilm forming ability; however, the biofilm-forming ability of some isolates in the absence of icaA gene highlights the importance of further genetic investigations of ica-independent biofilm formation mechanisms.
Keywords
Biofilm
icaA gene
multidrug resistance
staphylococci
Introduction
Conjunctival infections cause a worldwide problem and affect people of different ages. Proper treatment can reduce symptoms, recovery time, contagious spread, possible reinfection, and risk of complications but prolonged use of antibiotics can cause resistance strains. Infections with multidrug-resistant (MDR) organisms can lead to inadequate or delayed antimicrobial therapy.[1]
Staphylococcus spp. is a clinically relevant pathogen due to its antimicrobial resistance and evasion of the host immune system. Its virulence factors in avoiding host responses and influencing disease make them able to form biofilm. Emergence of resistant staphylococci from the conjunctiva is of great concern because its virulence is related to the clinical outcome of ocular infections such as keratitis or endophthalmitis.[2]
Biofilm-forming MDR Staphylococcus spp. are major reservoirs for transmission of ophthalmic infections. The ability of bacteria to aggregate and form biofilm is strictly related to the capacity of producing an extracellular mucoid substance: the slime, whose main component is of polysaccharide nature and consists of glycosaminoglycans.[3] Development of biofilm is considered to be a two-step process; first, the bacteria adhere to a surface mediated by a capsular antigen, namely, capsular polysaccharide/adhesin (PS/A), second, the bacteria multiply to form a multilayered biofilm, associated with production of polysaccharide intercellular adhesin (PIA) which mediates cell to cell adhesion.[4] It has been shown that both Staph aureus and S. epidermidis contain the intercellular adhesion (ica) operon responsible for slime production. This operon contains the intercellular adhesion gene A (icaA) DBC genes, in addition to the icaR gene which exerts a regulatory function. Among ica genes, the icaA and icaD have been reported to play a significant role in biofilm formation.[3]
The icaA gene responsible for PIA poly-N-succinyl β-1-6 glucosamine formation plays an independent role in the biofilm composition and intercellular adhesion. Staphylococcus spp., especially S. aureus, can produce a multilayered biofilm embedded within a glycocalyx or slime layer with heterogeneous protein expression throughout.[56]
The aim of this study is to determine association of icaA with biofilm formation in staphylococci isolated from patients with conjunctivitis, determine association of icaA gene with antibiotic resistance profile in staphylococci causing conjunctivitis, and to assess the reliability of two phenotypic methods used for biofilm detection.
Materials and Methods
Isolates
A total of 50 staphylococcal strains were obtained from 100 conjunctival swabs from patients complaining of inflammatory reactions in the eye (conjunctivitis) without or with other eye diseases or interventions attended the ophthalmology outpatient department of the Research Institute of ophthalmology; the study was conducted in the period from January 2015 to March 2016 who were diagnosed as staphylococcal species bacterial infection. The study protocol was approved by the Faculty of Medicine Ain Shams University Ethical Committee.
Specimen collection
Conjunctival swabbing was taken from the lower fornix of each case and was cultured onto surface of blood agar and mannitol salt agar plates (Oxoid, UK). Inoculated media were incubated aerobically at 37°C for maximum up to 48 h. Identification of Staphylococcus isolates was done by conventional methods including hemolytic activity on blood agar, acid production from mannitol, Gram-positive cocci in clusters by light microscopy, and catalase-positive reaction.[7] Final identification was done using Dry spot STAPHYTECT PLUS (Oxoid, UK) which is a latex slide agglutination test card used in the differentiation of Staphylococcus aureus by detection of clumping factor, Protein A, and certain polysaccharides.
Antimicrobial susceptibility using disc diffusion method
Antibiotic susceptibility of all isolates was done by disc diffusion method and results' interpretation was done according to.[8] Using a sterile loop, 3–5 well-isolated colonies from the pure culture of the organism were emulsified in sterile saline; the turbidity of the suspension was matched to the turbidity of 0.5 McFarland standards (BioMérieux, France) which was then swabbed on Muller Hinton agar (Oxoid, UK) supplemented with 2% NaCl. Antibiotics tested include penicillin, amoxicillin, ampicillin, cefoxitin, ceftriaxone, ciprofloxacin, vancomycin, amikacin, gentamycin, erythromycin, clindamycin, rifampicin, trimethoprim/sulfamethoxazole, and linezolid. Multidrug resistance was reported as a single isolate resistant (intermediate or complete) to three or more unique antimicrobial classes.[9]
Evaluation of biofilm formation by Staphylococcus isolates
Congo red agar (CRA method)
According to Freeman et al.,[10] the congo red agar (CRA) medium was prepared with 37 g/l BHI broth, 50 g/l sucrose, 10 g/l agar, and 0.8 g/l congo red. Congo red stain was prepared as a concentrated aqueous solution and autoclaved at 121°C for 15 min separately from other medium constituents and was then added when the agar had cooled to 55°C. Plates were inoculated and incubated at 37°C for 24 h. The plates were inspected for the color of the colonies at 24 and 48 h. A positive result was indicated by black colonies whereas nonproducing strains developed red colonies. The congo red dye directly interacts with certain polysaccharides, forming colored complexes or more likely some metabolic changes of the dye to form a secondary product which could play a more important part in the formation of dark colonies.[11] For colonies color evaluation, a four-color reference scale was used according to Satorres and Alcaráz:[12] black and bordeaux almost black were classified as biofilm producers, while bordeaux and red as nonbiofilm-producing strains. This method was performed in triplicate.
Microtiter plate method
Quantitative determination of biofilm production was carried by MTP according to Stepanović et al.[13] Staphylococcal strains were grown overnight at 37°C in BHI supplemented with 2% sucrose. The culture was adjusted to 0.5 McFarland then diluted 1:100 in fresh medium. Individual wells of sterile, polystyrene, 96-well, flat-bottomed tissue culture plates (TPP – Switzerland) were inoculated with 200 μl of the diluted culture then incubated. After 24 h incubation at 37 °C, the contents of each well were removed by decantation and each well was washed three times with 300 μl of phosphate-buffered saline (to remove free-floating “planktonic” bacteria). Biofilms formed by adherent ''sessile'' organisms were heat fixed by exposing them to hot air at 60 _C for 60 min, then 150 μl crystal violet (2%) stain was added to each well. After 15 min, the excess stain was rinsed off by decantation and the plate was washed and left to dry. Quantitative analysis of biofilm production was performed by adding 150 ll 95% ethanol to each well, and after 30 min, the optical densities (OD) of stained adherent bacterial films were read using a microtiter-plate reader (Tecan. Infinite F50) at 570 nm. Each assay was performed in triplicate. As a negative control, uninoculated medium was used to determine background OD. The average OD values were calculated for all tested strains and negative controls; the cutoff value (ODc) was established. It is defined as a three standard deviations (SD) above the mean OD of the negative control: ODc = average OD of negative control + (3 SD of negative control). ODc value was calculated for each microtiter plate separately. When a negative value was obtained, it was presented as zero, while any positive value indicated biofilm production. For interpretation of the results, strains were divided into the following categories:
-
Non biofilm producers = OD≤ODc
-
Weak biofilm producers = ODc<OD≤ 2XODc
-
Moderate biofilm producers = 2xODc<OD≤ 4xODc
-
Strong biofilm producers = 4xODc<OD.
Detection of intercellular adhesion gene A gene
DNA extraction
It was done using Qiagen DNeasy (Qiagen, USA), for DNA extraction from bacterial cultures according to manufacturer's instructions[13] (QIAgen DNeasy handbook, July, 2006).
Amplification of intercellular adhesion gene A gene by polymerase chain reaction
DNA from strong biofilm producing isolates was screened for icaA gene by polymerase chain reaction (PCR) in thermal cycler (Eppendorf, Hamburg, Germany).[14] Primers used were forward: AAGTCATACACTTGCTGGCG and reverse: CTGTCTGGGCTTCACCATGT. Reaction mixtures (50 μl) contained 25 μl PCR master mixtures, 1 μl of each primer (0.1–0.5 μM final concentration), 18 μl RNase free water, and 5 μl of template DNA. Amplifications were performed with the following thermal cycling profile; an initial denaturation at 94°C for 2 min, followed by 30 cycles of amplification (denaturation at 94°C for 1 min, primer annealing at 60°C for 1 min, and extension at 72°C for 2 min) and a final extension for 4 min. Amplicons for icaA-produced fragments of 188 bp were detected by Gel electrophoresis. The amplified product sizes were estimated by comparison with 100 bp DNA ladder (QIAGEN Incorporation) [Figure 1].
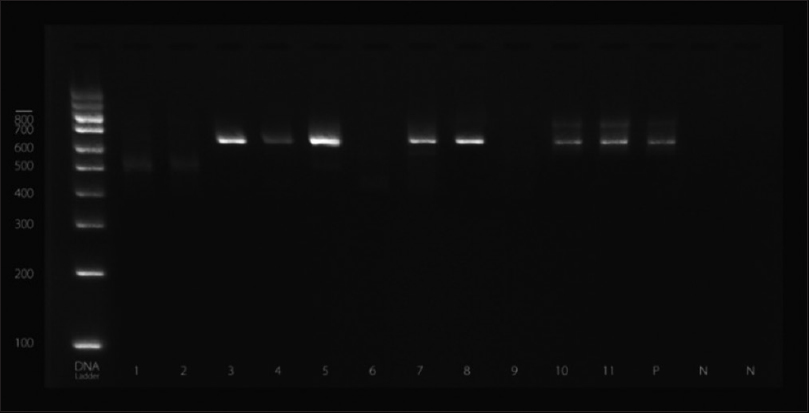
- Gel electrophoresis indicating the presence of intercellular adhesion gene A gene
Statistical analysis
Qualitative data were presented as frequencies (n) and percentages (%). Friedman's test and Wilcoxon signed-rank test were used to study the associations between the different modalities.
The significant level was set at P ≤ 0.05. Statistical analysis was performed with IBM® SPSS® Statistic Version 20 (IBM, USA, Washington).
Results
The study was conducted on 50 staphylococcal isolates; 30 S. aureus (60%) and 20 (40%) coagulase-negative staphylococci (CoNS).
Concerning detection of biofilm formation in S. aureus isolates by both CRA and MTP methods, 18 (60%) isolates were positive biofilm forming while 12 (40%) were negative. There was a statistically significant association between results of MTP and CRA. All the isolates (n = 18) detected as positive biofilm-forming S. aureus by CRA were also positive by MTP. Furthermore, all negative isolates (n = 12) detected by CRA were also negative by MTP. However, two isolates were different by both methods in the degree of biofilm formation [Table 1].
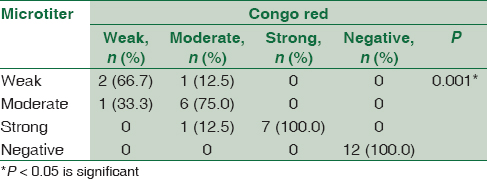
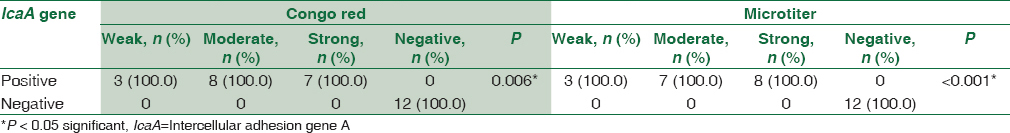
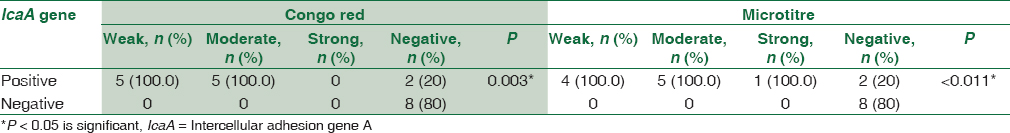
There was a statistically significant association between biofilm formation as detected by both MTP and CRA methods and the presence of icaA gene in S. aureus isolates as detected by PCR method. All weak, moderate, and strong cases as diagnosed by MTP and CRA were positive by PCR. All negative cases were negative by PCR [Table 2].
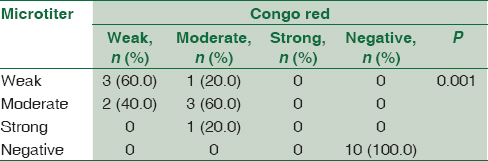
Concerning detection of biofilm formation in CoNS isolates by both CRA and MTP methods, 10 (50%) isolates were positive biofilm forming and 10 (50%) were negative; there was statistically significant association between results of MTP and CRA. All CoNS isolates (n = 10) that were negative for biofilm formation by CRA were also negative by MTP and all isolates (n = 10) that were positive by CRA were positive by MTP methods. However, three isolates were different by both methods in the degree of biofilm formation [Table 3].
There was a statistically significant association between biofilm formation as detected by both MTP and CRA and the presence of icaA gene in CoNS isolates as detected by PCR method. All weak, moderate, and strong cases as diagnosed by MTP and CRA were positive for icaA gene by PCR. All negative cases by CRA and MTP were negative for icaA except two [Table 4].
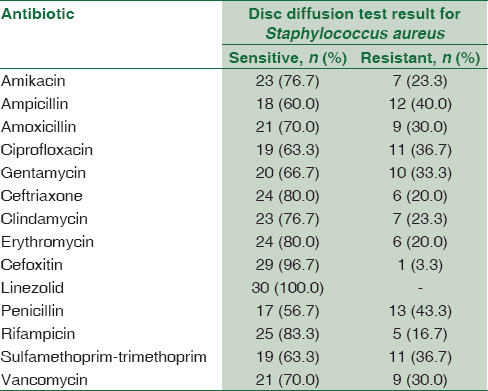
Concerning the result of antimicrobial susceptibility test, S. aureus isolates showed (100%) sensitivity to linezolid followed by cefoxitin (96.7%) then rifampicin (83.3%), ceftriaxone (80.7%), and erythromycin (80%); the least sensitivity was to penicillin (56.7%) and ampicillin (60%) [Table 5].
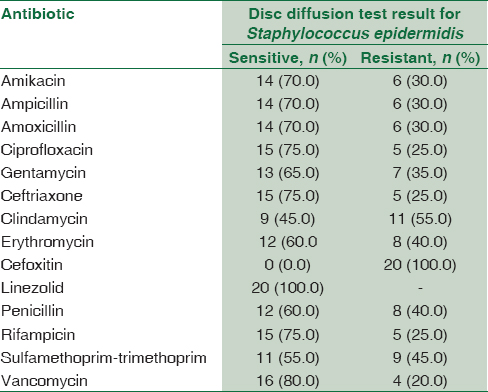
Concerning the result of antimicrobial susceptibility test, CoNS isolates showed (100%) sensitivity to linezolid followed by vancomycin (80%) then ceftriaxone, ciprofloxacin, and rifampicin (75%); the least sensitivity was to cefoxitin (0%), clindamycin (45%), and sulfamethoprim-trimethoprim (55%) [Table 6].
Concerning antibiotic susceptibility testing, this study shows that out of 30 S. aureus isolates, there were 19 (63.3%) isolates MDR to antibiotics, while 11 (55%) out of 20 isolates CoNS were MDR. By making correlation between the existence of icaA gene and MDR isolates, there was a statistically significant association between the presence of icaA gene and MDR in both S. aureus and CoNS. There was 89.5% of the MDR S. aureus positive for icaA gene and 72.7% of MDR CoNS were positive for icaA gene [Table 7].
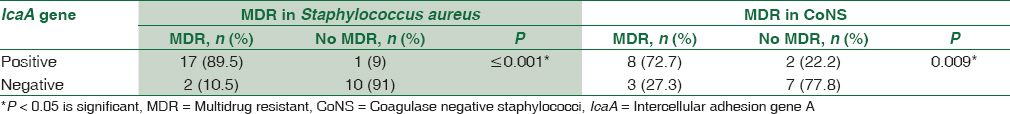
Discussion
S. aureus and CoNS are one of the frequent causes of bacterial conjunctivitis which involves biofilm formation.[14] One of the major virulence factors of S. aureus and CoNS is the slime production.[15] These bacteria have been known as a main cause of nosocomial infections. It is known that slime formation will significantly enhance the pathogenesis of staphylococcal infections. Furthermore, the slime formation allows staphylococci to escape host defenses and resists the antimicrobial action of antibiotics. Detection of slime production in staphylococcal strains isolated from clinical samples is important. It helps in discovering virulence factors of bacterial pathogenicity and hence the use of suitable ways to overcome them.[16]
It has been shown that both S. aurues and CoNS contain the ica operon responsible for slime production.[17]
The results of the present study revealed that biofilm production assessed by CRA revealed that 60% isolates of S. aureus were biofilm positive and 50% of CoNS were biofilm forming. In the present study, concerning detection of biofilm formation in S. aureus by both CRA and MTP methods, there was a highly significant association between the results of the two methods. All the isolates (n = 12) detected as negative biofilm-forming staphylococci by CRA method were also negative by MTP method. Furthermore, all positive biofilm-forming staph (n = 18) detected by CRA were also positive by MTP method. However, three of the biofilm forming S. aureus were different by both methods in the degree of formation of biofilm. One was detected as weak biofilm forming by CRA while detected by MTP as moderate biofilm forming. Two were detected as moderate biofilm-forming bacteria by CRA while detected by MTP as one weak and the other strong biofilm-forming S. aureus.
While observing the results concerning CoNS, this study revealed statistical association between the results of CRA and MTP in detecting biofilm formation. All CoNS isolates (n = 10) that were considered negative for biofilm formation by CRA method were also negative by MTP, and all the positive biofilm forming CoNS isolates (n = 10) were considered positive by both methods. However, CRA detected two of the isolates as weak compared to MTP method, which detect them as moderate biofilm-forming bacteria. Another two isolates were considered moderate biofilm forming by CRA while one was detected as weak and the other as strong biofilm-forming bacteria by MTP methods.
The previous results showed the high biofilm-forming ability of S. aureus and CoNS in conjunctival infections which was confirmed by both phenotypic methods of biofilm detection. Furthermore, the close results of CRA method and MTP reveal that we can use either method as a screening test for biofilm-forming ability in these organisms. These results agreed with those detected by Gad et al.[18] whose study showed high biofilm-forming ability (83.3%) in both S. aureus and CoNS isolated from urinary tract catheterized patients. Moreover, the study of Murgan et al., showed that 88.6% isolates of staphylococcal isolates from conjunctivitis was biofilm forming. Furthermore,[2021] showed high percentage high percentage of biofilm formation in CoNS reaching 63% and 71.4%, respectively,[22] suggested that phenotypic methods (the CRA method and the microtiter plate assay) could be used as putative screening methods for the detection of biofilm-positive strains of S. aureus and CoNS and may provide a convenient way for early diagnosis of biofilm-related infections.
As regards the result of antimicrobial susceptibility test, S. aureus isolates showed (100%) sensitivity to linezolid followed by cefoxitin (96.7%) then rifampicin (83.3%), ceftriaxone (80.7%), and erythromycin (80%); the least sensitivity was to penicillin (56.7%) and ampicillin (60%) comparing to the result of[23] who reported that 31% of S. aureus were multidrug resistance (MDR) and 42.7% were oxacillin resistant. Most of these strains were isolated from wound specimens. All isolates were susceptible to vancomycin (100%). They were also susceptible to chloramphenicol, linezolid, nitrofurantoin, rifampicin, and teicoplanin (>80%) but showed resistance to erythromycin and penicillin.
Concerning the result of antimicrobial susceptibility test in CoNS isolates showed (100%) sensitivity to linezolid followed by vancomycin (80%) then ceftriaxone, ciprofloxacin, and rifampicin (75%); the least sensitivity was to cefoxitin (0%), clindamycin (45%), and sulfamethoprim-trimethoprim (55%) compared to the result of Al Tayyar et al.[24] who reported that antibiotic susceptibility testing of CoNS species revealed their sensitivity to vancomycin, linezolid, rifampin, and nitrofurantoin, while showing a highly resistant pattern to ampicillin, penicillin, ceftriaxone, cefazolin, amoxicillin-clavulanic acid, and erythromycin. Moreover, the study of Murad et al.[25] reported that antimicrobial resistance of CoNS were oxacillin (70.3%), amoxicillin (74.8%), amoxicillin + clavulanate (32.8%), ciprofloxacin (35.2%), ofloxacin (33.6%), ceftriaxone (30.4%), erythromycin (58.3%), clindamycin (16.3%), daptomycin (42.5%), kanamycin (52.2%), fusidic acid (41.7%), doxycycline (24.7%), vancomycin (2.6%), and linezolid (0.8%).
This study shows high association between biofilm formation and icaA gene presence in S. aureus isolates; ica A gene was positive in all the weak, moderate, and strong biofilm-forming S. aureus isolates (100%), and all the negative biofilm-forming S. aureus (100%) were also observed negative for icaA gene presence. In CoNS isolates, it was also observed that all weak, moderate, and strong biofilm-forming isolates as diagnosed by MTP and CRA were positive for icaA gene by PCR. All negative cases by CRA and MTP were negative for icaA except two. These results agreed with those obtained by Gad et al.[18] who found that all biofilm-producing strains of S. aureus and CoNS from urinary tract catheterized patients were positive for the icaA gene, while the biofilm-negative strains were negative for the gene. Freeman et al.[10] and Murugan et al.[19] reported that all S. aureus and CoNS biofilm-positive strains isolated from intravenous catheters and eye infections were positive for icaA gene and that this gene is required for full slime synthesis, which is in agreement with the results of this study. In addition, in the study of Arciola et al.,[11] only one out of 65 staphylococci was found to be biofilm negative by CRA while possessing the icaA and icaD genes.
Some authors attributed the absence of biofilm production in some staphylococcal isolates despite the presence of the ica operon to the insertion of a 1332-bp sequence element, known as IS256, in icaA causing its inactivation.[26] However, the transposition of IS256 into the ica operon has been found to be a reversible process as after repeated passages of the PIA-negative insertional mutants, the biofilm-forming phenotype could be restored. Cafiso et al.[27] also proposed that the product of icaR gene (a regulator gene which seems to function as a repressor) could influence transcription of the ica operon. Nevertheless, irrespective of ica genes expression, ica positive isolates should be considered to be potential biofilm producers.[17]
In this study, there was a high significant association between icaA gene presence in Staphylococcus strains and resistance to various antibiotics. icaA gene was positive in 89.5% of the MDR S. aureus and 72.7% of MDR CoNS. These results are greatly consistent with those obtained by Murugan et al.[19] who reported that all the MDR staphylococcal strains isolated from conjunctival infections carry the icaA gene; Abdolmajid et al.[28] also reported in their study that 72% of the isolated MDR staph strains carry the ica operon which agreed with this study.
From the previous results, we can conclude the great correlation between biofilm formation and icaA gene presence in staph spp. Furthermore, there is a great correlation between MDR Staphylococcal spp. and icaA gene carriage. MDR isolates of Staphylococcal spp. usually carry icaA gene which is capable of strong biofilm formation that contributes to the higher antibiotic resistance.
Conclusion
The icaA gene responsible for slime production among the Staphylococcal isolated from conjunctival infection has been found in a significantly high frequency. On the other hand, the biofilm-forming ability of some strains in the absence of icaA gene highlights the importance of further genetic investigations of ica-independent biofilm formation mechanisms. A suitable and reproducible method is necessary for screening of biofilm producers in any healthcare setup, and CRA test is recommended as it is easier to perform, cheap, and the agreement between the CRA plate test with the molecular detection of ica genes indicates that it is a reliable test. Further investigations on inhibiting icaA gene are recommended as a gene responsible for biofilm formation may minimize the selection for antimicrobial resistance strains.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Future of ophthalmic anti-infective therapy and the role of moxifloxacin ophthalmic solution 0.5% (VIGAMOX) Surv Ophthalmol. 2005;50(Suppl 1):S64-7.
- [Google Scholar]
- Bacterial susceptibility to topical antimicrobials and clinical outcome in bacterial keratitis. Invest Ophthalmol Vis Sci. 2010;51:362-8.
- [Google Scholar]
- Detection of biofilm-forming strains of Staphylococcus epidermidis and S. aureus. Expert Rev Mol Diagn. 2002;2:478-84.
- [Google Scholar]
- Detection of icaAD gene and biofilm form in Staphylococcus aureus isolates from wound infections. Iran Public Health. 2006;35:25-8.
- [Google Scholar]
- The gene bap, involved in biofilm production, is present in Staphylococcus spp. Strains from nosocomial infections. J Microbiol. 2009;47:319-26.
- [Google Scholar]
- Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888-96.
- [Google Scholar]
- District Laboratory Practice in Tropical Countries. Norfolk: Cambridge University Press; 2006. p. :434-40.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. In: CLSI Document M100-s20. Wayne, PA: CLSI; 2016.
- [Google Scholar]
- Multidrug resistance: An emerging crisis. Interdiscip Perspect Infect Dis 2014 2014:541340.
- [Google Scholar]
- New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872-4.
- [Google Scholar]
- Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39:2151-6.
- [Google Scholar]
- Prevalence of icaA and icaD genes in Staphylococcus aureus and Staphylococcus epidermidis strains isolated from patients and hospital staff. Cent Eur J Public Health. 2007;15:87-90.
- [Google Scholar]
- Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891-9.
- [Google Scholar]
- Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet Microbiol. 2003;92:179-85.
- [Google Scholar]
- Detection of adhesin genes and slime production among staphylococci in orthopaedic surgical wounds. Afr J Microbiol Res. 2010;4:708-15.
- [Google Scholar]
- Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res Notes. 2010;3:260.
- [Google Scholar]
- Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. J Infect Dev Ctries. 2009;3:342-51.
- [Google Scholar]
- Biofilm forming multi drug resistant Staphylococcus spp. Among patients with conjunctivitis. Pol J Microbiol. 2010;59:233-9.
- [Google Scholar]
- Isolation, vancomycin resistance and biofilm production of Staphylococcus epidermidis from patients with conjunctivitis, corneal ulcers, and endophthalmitis. Rev Latinoam Microbiol. 2006;48:238-46.
- [Google Scholar]
- Slime-producing Staphylococcus epidermidis and S. aureus in acute bacterial conjunctivitis in soft contact lens wearers. New Microbiol. 2005;28:345-54.
- [Google Scholar]
- Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:491-510.
- [Google Scholar]
- Antimicrobial susceptibility pattern of Staphylococcus aureus isolated from clinical specimens in Northern area of Jordan. Iran J Microbiol. 2015;7:265-72.
- [Google Scholar]
- Prevalence and antimicrobial susceptibility pattern of coagulase-negative staphylococci (CoNS) isolated from clinical specimens in Northern of Jordan. Iran J Microbiol. 2015;7:294-301.
- [Google Scholar]
- Identification and antibiotic susceptibility pattern of coagulase-negative Staphylococci in various clinical specimens. Park J Med Sci. 2013;29:1420-4.
- [Google Scholar]
- Detection of the icaADBC gene cluster and biofilm formation in Staphylococcus epidermidis isolates from catheter-related urinary tract infections. Int J Antimicrob Agents. 2002;19:570-5.
- [Google Scholar]
- Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004;10:1081-8.
- [Google Scholar]
- Biofilm formation in multi-drug resistant Staphylococcus aureus isolates from hospitalized patients. J Infect Dis. 2014;1:2411-20.
- [Google Scholar]





