Translate this page into:
Detection of VIM and NDM-1 metallo-beta-lactamase genes in carbapenem-resistant Pseudomonas aeruginosa clinical strains in Bahrain
Address for correspondence: Dr. Ronni Mol Joji, Department of Microbiology, Immunology and Infectious Disease, College of Medicine and Medical Sciences, Arabian Gulf University, Manama, Kingdom of Bahrain. E-mail: ronnimj@agu.edu.bh
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
INTRODUCTION:
Carbapenem-resistant Pseudomonas aeruginosa has emerged as a life-threatening infectious agent worldwide. Carbapenemase genes are reported to be some of the most common mechanisms for carbapenem resistance in P. aeruginosa. No reports are available from the Kingdom of Bahrain about carbapenem resistance and the underlying cause. In this study, we determined to study the presence of the metallo-beta-lactamase (MβL) genes of VIM family and NDM-1 in carbapenem-resistant P. aeruginosa strains.
METHODOLOGY:
Fifty carbapenem-resistant P. aeruginosa isolates were obtained from three main hospitals of Bahrain. They were subjected to antimicrobial susceptibility testing by disc diffusion test. Subsequently, MβL was detected by imipenem-ethylene diamine tetraacetic acid (EDTA) combined disc test and conventional polymerase chain reaction.
RESULTS:
Among 50 P. aeruginosa strains, 40 (80%) were imipenem resistant. Among the 40 imipenem-resistant strains, 35 (87.5%) strains were positive for the imipenem-EDTA combined disc test, and 21 (52%) were carrying MβL genes. Nineteen (47.5%) strains were positive for the VIM gene; one (2.5%) strain was carrying the NDM-1 gene, while one strain was carrying both the VIM and NDM-1 genes. None of the imipenem sensitive strains carried the VIM or NDM-1 gene.
CONCLUSION:
This is the first study to report the presence of the VIM family gene and NDM-1 genes in imipenem-resistant P. aeruginosa isolates in the Kingdom of Bahrain. The study also confirms the multiple drug resistance by the MβL strains, attention should therefore from now on, be focused on prevention of further spread of such isolates by firm infection control measures, and to reduce its threat to public health.
Keywords
Carbapenem resistance
NDM-1 gene
Pseudomonas aeruginosa
VIM gene
Introduction
Carbapenem resistance among Pseudomonas aeruginosa is a global health threat and has led to therapeutic limitations.[1] Carbapenemase genes which code for carbapenemases is reported to be an important mechanism in carbapenem resistance of P. aeruginosa.[2]
Carbapenemases are assigned to three classes of β lactamases: Ambler A, B, and D.[3] Class B metallo-beta-lactamases (MβL) include the enzymes that belong to VIM, IMP, SPM, GIM, and NDM families.[1] They hydrolyze all β-lactams, except aztreonam, and this activity can be inhibited by ethylene diamine tetraacetic acid (EDTA).[4] Nothing is known about P. aeruginosa MβL producers in the Kingdom of Bahrain.
The most relevant epidemiologically and clinically important MβL types are VIM (Verona integrin-encoded MβL), IMP (imipenemase), NDM (New Delhi MβL), and SPM (Sao Paulo MβL).[56]
Here, we studied the VIM family and NDM-1 genes among MβL-producing P. aeruginosa isolates by phenotypic test and polymerase chain reaction (PCR).
Methodology
Strain collection
The study was conducted following ethical approval from the Ethical Review Board of Arabian Gulf University (E014-PI-11/16). The study was conducted on 50 nonduplicate P. aeruginosa strains isolated from clinical samples from 50 patients (included community, wards, and ICU patients) attending the Salmaniya Medical Complex, King Hamad University Hospital, and Bahrain Defense Force Hospital, located in the Kingdom of Bahrain. The isolates were preserved in 20% skimmed milk with glycerol solution and stored in a freezer at −80°C until further processing.
Inclusion criteria
This study included all the P. aeruginosa strains isolated from patients of all the age groups and both the sexes. This included both the outpatients and the inpatients attending all the three hospitals in Bahrain.
Exclusion criteria
Repeat isolates from the same patients were excluded from the study.
The samples were collected under complete aseptic conditions and included wound swabs, sputum, deep tracheal aspirates, endotracheal tube, urine, blood, and tissue.
Identification of Pseudomonas aeruginosa
The isolates were identified as P. aeruginosa strains by standard laboratory techniques such as Gram staining, colony morphology, cetrimide test, catalase test, oxidase reaction, citrate utilization, TSI reaction, oxidation-fermentation test, gelatin hydrolysis test, polymyxin B sensitivity testing, and sugar fermentation tests.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by the disc diffusion method on Mueller-Hinton agar (MHA) plates and interpreted according to Clinical Laboratory Standards Institute recommendations (CLSI 2016). The antibiotic discs used were as follows: imipenem (10 μg), meropenem (10 μg), amikacin (30 μg), gentamycin (10 μg), ceftazidime (30 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), piperacillin + tazobactum (100/10 μg), tigecycline (15 μg), and colistin (10 μg).
Phenotypic detection of metallo-beta-lactamase activity
All the isolates resistant to imipenem (zone size ≤15 mm as per the CLSI guidelines 2016) by disc diffusion method on MHA were screened for MβL activity by imipenem-EDTA combined disc test (IMP-EDTA CDT) as described by Yong et al.[7] In brief, an overnight culture of the test organism was compared with 0.5 McFarland which is comparable to the density of bacterial suspension 1.5 × 108 CFU/ml and was inoculated on an MHA plate. Two imipenem discs (10 μg) were placed on the inoculated plate at a distance of 5 cm from each other, and 10 μl of 0.5M EDTA solution was added to one of the discs. The plates were incubated at 35°C for 16–18 h. The inhibition zones of each disc were compared, and the test isolates which showed a zone size of ≥7 mm for IMI-EDTA disc as compared to imipenem disc alone were considered as MβL positive [Figure 1]. P. aeruginosa ATCC 27853 strain was used as the control strain.
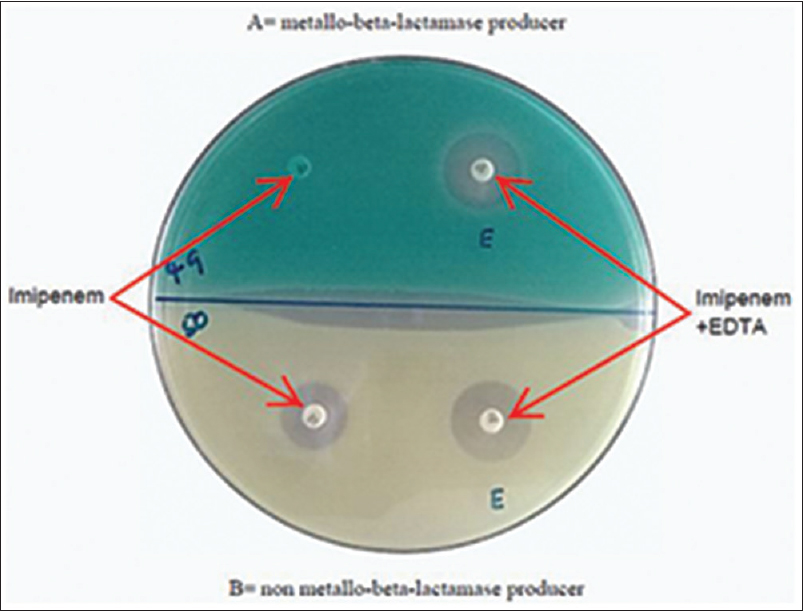
- Photo of a Combined Disc Test showing strain A as metallo-beta-lactamase producer and strain B as non metallo-beta-lactamase producer
Polymerase chain reaction assay for detection of metallo-beta-lactamase genes of VIM family and NDM-1
Conventional PCR testing of all the isolates for the presence of the VIM family and NDM-1 genes was done as per manufacturer's instructions. The sequence of primers specific for VIM family and NDM-1 used in this study is listed in Table 1.[8] Total DNA of all the bacterial isolates was extracted by the boiling method.[9] The extracted DNA was then stored at −20°C until further processing. Amplification was done using GoTaq Green PCR Master Mix (Promega). The PCR mix consisted of 25 μl master mix, 1 μl each of forward and reverse primer, 5 μl template DNA and nuclease-free water to make a final volume of 50 μl. The thermal cycler program was as follows: initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 30 s, followed by final extension at 72°C for 10 min. Agarose gel electrophoresis for the detection of amplicons was done by separating 10 μl of each amplicon and 100 bp ladder on a 1.5% agarose gel. Amplicons were visualized using ultraviolet transilluminator and subsequently analyzed.
| Gene | Primer sequence (5’- 3’) | Amplicon size |
|---|---|---|
| VIM | Vim-F GAT GGT GTT TGG TCG CAT A | 390 bp |
| Vim-R CGA ATG CGC AGC ACC AG | ||
| NDM-1 | NDM-1 F CAT TAG CCG CTG CAT TGA TG | 445 bp |
| NDM-1 R GCG AAA GTC AGG CTG TGT TG |
Statistical analysis
Data were analyzed using the Statistical Package for the Social Science (SPSS) version 20 (IBM, Armonk, NY, United States of America) to obtain descriptive data. Kappa test was used to measure the level of agreement between the phenotypic and genotypic test.
Results
The majority of the Pseudomonas aeruginosa strains are imipenem resistant
Fifty nonduplicate P. aeruginosa strains were isolated from clinical sources. The antimicrobial susceptibility test of these samples shows that out of 50 P. aeruginosa strains, 40 (80%) were imipenem resistant. All the isolates were resistant to ciprofloxacin (100%). 90% of the strains were resistant to norfloxacin, meropenem, and piperacillin/tazobactam which is shown in Table 2.
| Antibiotic | Number of resistant isolates (%) |
|---|---|
| Ciprofloxacin | 50 (100) |
| Norfloxacin | 45 (90) |
| Meropenem | 45 (90) |
| Imipenem | 40 (80) |
| Ceftazidime | 43 (86) |
| Cefotaxime | 43 (86) |
| Tigecycline | 38 (76) |
| Piperacillin/tazobactam | 45 (90) |
| Gentamicin | 43 (86) |
| Amikacin | 36 (72) |
| Colistin | 0 |
Most of the imipenem-resistant strains are positive for metallo-beta-lactamase
To determine whether imipenem resistance is caused by the production of MβL or by other mechanisms, the 40 imipenem-resistant strains were analyzed with the imipenem-EDTA combined disc test. An example of the imipenem-EDTA combined disc test is shown in Figure 1. This phenotyping revealed that 35 (88%) of these 40 strains were positive for MβL.
More than half of the imipenem resistance is due to VIM family and NDM-1 genes
Genotyping of all 50 strains was performed to determine the presence of the VIM family and NDM-1 genes. In Figures 2 and 3, the analyses of the PCR products are illustrated for the VIM and NDM-1 genes, respectively. The results showed that of the 40 imipenem-resistant strains, 19 (47.5%) were positive for the VIM gene, one isolate (2.5%) for the NDM-1 gene, and one (2.5%) was carrying both the VIM and NDM-1 genes, as shown in Table 3. None of the imipenem sensitive strains were carrying these genes.
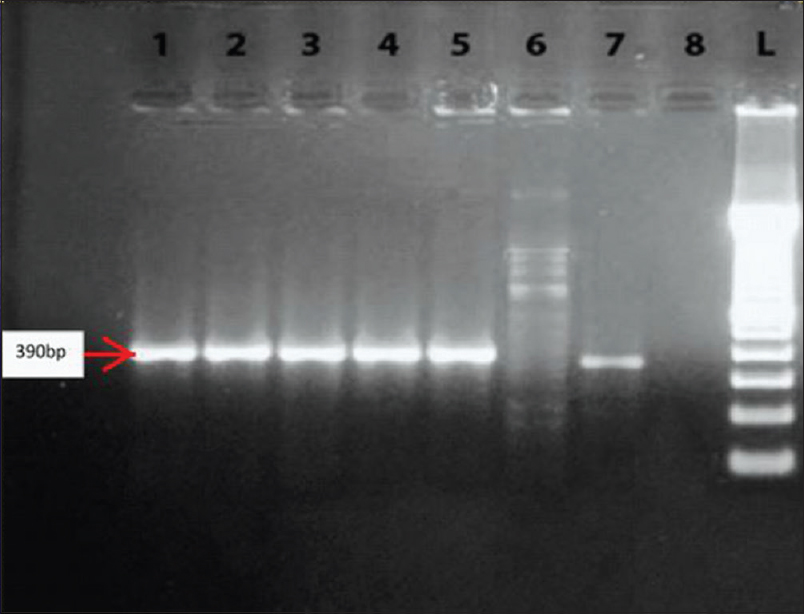
- Polymerase chain reaction products after agarose gel electrophoresis. Lanes 1-5 and 7 show one band with molecular size 390 bp (VIM gene), lane 6 is a VIM negative strain, and lane 8 is water (negative control). Lane L contains a 100 bp ladder
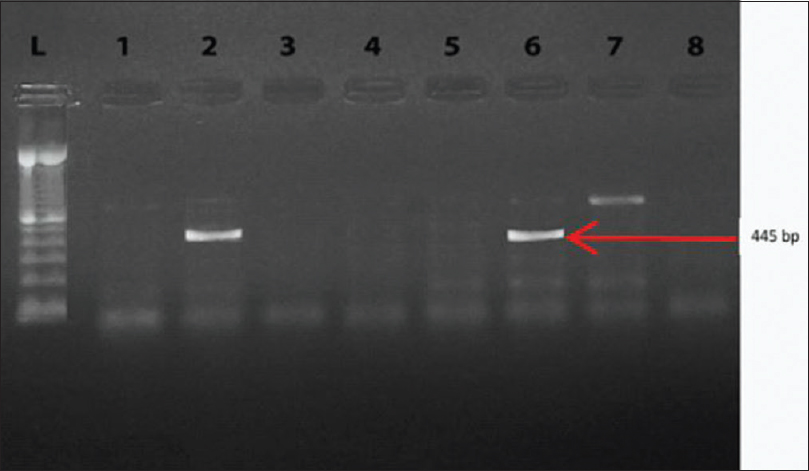
- Polymerase chain reaction products after agarose gel electrophoresis. Lanes 2 and 6 show one band with molecular size 445 bp (NDM-1 gene, lanes 1,3,4,5, and 7 are NDM-1 negative strains and lane 8 is water (negative control). Lane L contains a 100 bp ladder
| Imipenem sensitive, total isolates=10 | Imipenem resistant, total isolates=40 (%) | |
|---|---|---|
| VIM | 0 | 19 (47.5) |
| NDM-1 | 0 | 1 (2.5) |
| VIM + NDM-1 | 0 | 1 (2.5) |
| Total | 0 | 21 (52.5) |
Many of the VIM and NDM-1 gene-positive MβL producers were isolates obtained from endotracheal aspirate (seven of 19 VIM-positive strains [37%] and one of two NDM-1-positive strains [50%]) as can be seen in Table 4. These MβL producers were also resistant to most of the other antibiotics tested while they were all sensitive to colistin [Table 5].
| Clinical isolate | VIM positive, total=19 (%) | NDM-1 positive, total=2 (%) |
|---|---|---|
| Endotracheal aspirate | 7 (37) | 1 (50) |
| Swab* | 5 (26) | 1 (50) |
| Blood | 1 (5) | |
| Respiratory secretions | 4 (21) | |
| Urine | 1 (5) | |
| Tissue | 1 (5) |
*One sample was positive for both VIM and NDM-1
| Antibiotic | MβL producers (total isolates=21) | ||
|---|---|---|---|
| Resistant (%) | Intermediate | Sensitive (%) | |
| Imipenem | 21 (100) | - | - |
| Meropenem | 18 (86) | - | 3 (14) |
| Amikacin | 14 (67) | - | 7 (33) |
| Gentamycin | 17 (81) | - | 4 (19) |
| Ceftazidime | 18 (86) | - | 3 (14) |
| Cefotaxime | 17 (81) | - | 4 (19) |
| Ciprofloxacin | 21 (100) | - | |
| Norfloxacin | 18 (86) | - | 3 (14) |
| Piperacillin + tazobactam | 17 (81) | - | 4 (19) |
| Tigecycline | 16 (76) | - | 5 (24) |
| Colistin | - | - | 21 (100) |
MβL = Metallo-beta-lactamase
Correlation of the phenotypic test results with polymerase chain reaction
When comparing the phenotypic test results with genotypic test results, we found that the strength of agreement was fair between the two tests (kappa value = 0.47) as shown in Table 6. The sensitivity and specificity of the combined disc test in relation to PCR was 100% and 51.72%, respectively.
| CDT | PCR | κ | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Positive | 21 | 14 | 35 | 0.47 |
| Negative | 0 | 15 | 15 | |
| Total | 21 | 29 | 50 | |
CDT = Combined disc test, PCR = Polymerase chain reaction
Discussion
P. aeruginosa is a multidrug-resistant organism causing nosocomial infections.[10] Over the past decades, the resistance to carbapenems is increasing and has become a major health threat.[11] Early detection of MβL producers is therefore very crucial for optimal treatment of infections, and this will further reduce the resistance rate and prevent nosocomial spread. In the present study, out of 50 P. aeruginosa isolates, 40 (80%) were found to be resistant to imipenem which is in agreement with the study by Polotto et al.[12] in Brazil where they reported 96.4% of their strains as imipenem resistant. Another study, in India, by Arunagiri et al.[13] reported 62.7% of the isolates as resistant to imipenem. In Egypt, EL-Mosallamy et al.[8] conducted a study on 100 strains, wherein they found 25 (25%) strains imipenem resistant. In Saudi Arabia, Mohamed et al.[14] reported that imipenem resistance rate was 38.6% in 2011, while 5 years later, another study reported that 91% of 33 P. aeruginosa isolates were resistant to imipenem.[15]
There are no official standard guidelines for MβL detection. PCR analysis is the gold standard, but it is not practiced in routine microbiology laboratories. We therefore first used IMI-EDTA CDT phenotyping for MβL screening and compared the results with the genotyping results. By IMI-EDTA CDT phenotyping, we observed that out of 40 imipenem-resistant strains, 35 (88%) produced MβL whereas Pitout et al.[16] from Canada found that 110/241 (46%) imipenem-resistant strains were MβL positive while in Iran, Saderi et al.[17] reported that 65/100 (65%) of their imipenem-resistant strains were MβL positive using phenotypic methods. Another study by Panchal et al. compared different phenotypic tests for MβL detection and found that 19/30 (63.33%) were positive by IMI-EDTA combined disc test.[18]
The PCR results revealed that out of 40 imipenem-resistant strains, 19 (47.5%) strains were positive for the VIM gene which is similar to a study from Egypt by Essa and Afif where they found 40% of their imipenem-resistant strains carrying the VIM gene.[18] Al-Agamy et al. from Saudi Arabia reported 20.6% of the imipenem-resistant strains as MβL producers and all the MβL strains were found to carry the VIM gene.[19] Furthermore, in a study by Tawfik et al. from Saudi Arabia, VIM was found in all the 15 MβL-positive isolates (100%).[20] In a study in Canada, 43% of the strains were positive for the VIM gene.[16]
Resistance transferred by the NDM-1 gene is also a growing public health problem. The main reservoir is the Indian subcontinent, and the secondary reservoirs are the Balkans regions and the Middle East.[21] Here, we observed only one isolate (2.5%) positive for NDM-1 gene which is in corroboration with a study from Egypt by Zafer et al. which concluded that the prevalence of the NDM-1 gene was only 4.2%.[22] Another study by Shanthi et al.[23] from India in 2014 reported that only four isolates out of 61 were positive for NDM-1. We observed only one isolate that carried both the VIM and NDM-1 genes, whereas in Saudi Arabia, Shaaban et al.[24] reported 8 out of 16 imipenem-resistant strains carrying both NDM-1 and VIM subtypes (VIM 1 and VIM 2). A few previous studies have also reported the presence of multiple carbapenemase genes in P. aeruginosa, including the KPC and VIM in Colombia[25] and SPM-1, KPC-2, and VIM-2 in Brazil.[2] The differences in the incidence and the types of genes seen in MβL producing strains are likely due to the geographical variations and differences in antibiotic usage.
The strength of agreement between the combined disc test and PCR is moderate. The sensitivity and specificity of IMI-EDTA CDT in relation to PCR is 100% and 51.72%, respectively, which was similar to the studies conducted by Pandya et al.[26] and Arunagiri et al.[13] where they reported sensitivity of IMI-EDTA CDT as 96.3% and 94%, respectively, while Picão et al.[27] reported a lower sensitivity of 80%.
Conclusion
This is the first study to report the presence of VIM and NDM-1 in imipenem-resistant P. aeruginosa strains in the kingdom of Bahrain. The test results also showed that imipenem-EDTA combined disc test is a sensitive method for the detection of MβL producers. This test can, therefore, be used as an alternative to PCR in diagnostic laboratories. The study also identified the multiple drug resistance of the MβL producers. Attention should be focused on early detection of MβL producers to prevent further spread of such multidrug-resistant strains. The development of strong antimicrobial stewardship programs is essential, with emphasis on the importance of infection control measures to prevent further spread of these strains.
Financial support and sponsorship
This study was supported by Arabian Gulf University.
Conflicts of interest
There are no conflicts of interest.
References
- Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at mulago hospital in Kampala, Uganda (2007-2009) Springerplus. 2016;5:1308.
- [Google Scholar]
- Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann Clin Microbiol Antimicrob. 2014;13:43.
- [Google Scholar]
- The spread of carbapenemase-producing bacteria in Africa: A systematic review. J Antimicrob Chemother. 2015;70:23-40.
- [Google Scholar]
- Evaluation of phenotypic tests for detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains in china. J Clin Microbiol. 2009;47:1136-42.
- [Google Scholar]
- Metallo-beta-lactamases: The quiet before the storm? Clin Microbiol Rev. 2005;18:306-25.
- [Google Scholar]
- Boronic acid disk diffusion for the phenotypic detection of polymerase chain reaction-confirmed, carbapenem-resistant, gram-negative bacilli isolates. BMC Microbiol. 2016;16:135.
- [Google Scholar]
- Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. And Acinetobacter spp. J Clin Microbiol. 2002;40:3798-801.
- [Google Scholar]
- Phenotypic and genotypic methods for detection of metallo-beta-lactamase (MBL) producing Pseudomonas aeruginosa. Egypt J Med Microbiol. 2015;24:27-35.
- [Google Scholar]
- Quality improvement of the DNA extracted by boiling method in gram negative bacteria. Int J Bioassays. 2017;6:5347-9.
- [Google Scholar]
- Nosocomial outbreak of imipenem-resistant Pseudomonas aeruginosa producing VIM-2 metallo-β-lactamase in a kidney transplantation unit. Diagn Pathol. 2011;6:106.
- [Google Scholar]
- Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582-610.
- [Google Scholar]
- Detection of P. Aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis. 2012;12:176.
- [Google Scholar]
- Detection and characterization of metallo-beta-lactamases in Pseudomonas aeruginosa by phenotypic and molecular methods from clinical samples in a tertiary care hospital. West Indian Med J. 2012;61:778-83.
- [Google Scholar]
- Antimicrobial resistance pattern and prevalence of metallo-β-lactamases in Pseudomonas aeruginosa from Saudi Arabia. Afr J Microbiol Res. 2011;5:5528-33.
- [Google Scholar]
- Diversity of molecular mechanisms conferring carbapenem resistance to Pseudomonas aeruginosa isolates from Saudi Arabia. Can J Infect Dis Med Microbiol. 2016;2016:4379686.
- [Google Scholar]
- Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129-35.
- [Google Scholar]
- Detection of Metallo-β-Lactamase Producing Pseudomonas aeruginosa Isolated From Burn Patients in Tehran, Iran. Lab Med. 2010;41:609-12.
- [Google Scholar]
- Comparison of four phenotypic methods for detection of metallo-β-lactamase-producing gram-negative bacteria in rural teaching hospital. J Lab Physicians. 2017;9:81-3.
- [Google Scholar]
- Antimicrobial resistance pattern and prevalence of MBL in P.aeruginosa from Saudi Arabia. Afr J Microbiol Res. 2011;30:5528-33.
- [Google Scholar]
- Distribution of ambler class A, B and D β-lactamases among Pseudomonas aeruginosa isolates. Burns. 2012;38:855-60.
- [Google Scholar]
- Worldwide dissemination of the NDM-type carbapenemases in gram-negative bacteria. Biomed Res Int. 2014;2014:249856.
- [Google Scholar]
- Antimicrobial resistance pattern and their beta-lactamase encoding genes among Pseudomonas aeruginosa strains isolated from cancer patients. Biomed Res Int. 2014;2014:101635.
- [Google Scholar]
- Detection of new delhi metallo beta lactamase-1 (NDM-1) carbapenemase in Pseudomonas aeruginosa in a single centre in southern india. Indian J Med Res. 2014;140:546-50.
- [Google Scholar]
- Molecular characterization of resistance mechanisms in Pseudomonas aeruginosa isolates resistant to carbapenems. J Infect Dev Ctries. 2017;11:935-43.
- [Google Scholar]
- First report of a Pseudomonas aeruginosa isolate coharboring KPC and VIM carbapenemases. Antimicrob Agents Chemother. 2012;56:5422-3.
- [Google Scholar]
- Evaluation of various methods for detection of (MβL) production in gram negative bacilli. Egypt J Med Microbiol. 2011;2:775-7.
- [Google Scholar]
- Metallo-beta-lactamase detection: Comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol. 2008;46:2028-37.
- [Google Scholar]





