Translate this page into:
Evaluation of performance of direct disk diffusion test from positively flagged blood culture broth: A large scale study from South India
Address for correspondence: Dr. Apurba S. Sastry, Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry ‑ 605 006, India. E-mail: drapurbasastry@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Rapid turnaround time of blood culture reports should be the main motive for a clinical microbiologist for optimal patient care. Categorical agreement (CA) between direct disk diffusion (dDD) and reference disk diffusion (rDD) may vary between laboratories.
AIMS AND OBJECTIVES:
This study was designed to determine the CA and understand various types of errors associated with antibiotic organism combination, so that caution can be derived while interpreting and reporting dDD results in the earliest meaningful time frame.
MATERIALS AND METHODS:
In the present study, dDD results were compared to the rDD results from the positive blood culture bottles. CA and various types of errors were evaluated.
RESULTS:
A total of 965 pathogens and 7106 organism antibiotic combinations were evaluated in this study. Overall, there was a CA of 96% which was extremely satisfactory. The categorical disagreement was found only in 4% of organism antibiotic combinations; majority of which were major error (ME, 2.1%) followed by very ME (1%) and minor error (0.9%). The errors were marginally high for Enterobacteriaceae testing against β lactam- β lactamase inhibitor combinations, for Pseudomonas species against aminoglycosides and ciprofloxacin and Staphylococcus species against cefoxitin, one should be vigilant while reporting dDD result of these antibiotic organism combinations.
CONCLUSION:
dDD is of paramount importance for early institution of targeted therapy and is considered as one of the key stewardship intervention. Our study gives an insight that every laboratory must perform dDD for positively flagged blood culture specimens; the result of which should be confirmed later by performing rDD. One should be vigilant while reporting dDD result of BL BLI for Enterobacteriaceae; aminoglycosides and CF for Pseudomonas species; cefoxitin for Staphylococcus species and HLG for Enterococcus species. Supplementary tests such as MRSA latex should be included when necessary.
Keywords
Blood culture
direct susceptibility test
disk diffusion
Introduction
Sepsis is one of the major causes of mortality and morbidity in hospitalized patients. Blood culture is the gold standard method for the diagnosis of sepsis, and it is included among the early investigation to be sent for sepsis according to the Surviving Sepsis Campaign guidelines.[1] There is an increase in mortality by about 7.6% with every hour of delay in the initiation of appropriate antimicrobial therapy.[2] Literature suggests that about 40% of all patients with bacteremia receive inadequate antibiotic treatment until the first notification of a positive blood culture.[3] Even with advancement in molecular diagnostics, for all practical purposes, blood culture still remains the most important microbiological investigation in the management of sepsis. Rapid turnaround time (TAT) of blood culture reports should be the main motive for a clinical microbiologist for optimal patient care. Collecting recommended volume of blood aseptically for blood culture, sending at least two sets of culture before administering the first dose of antibiotics, utilizing automated blood culture systems over conventional culture, initial reporting of Gram stain report of positive blood culture bottles are few of the measures in reducing TAT of blood culture reports.[45]
As per the standard practice in most of the blood culture laboratory, it takes around 48 h to report antibiotic sensitivity results after a blood culture bottle is flagged positive by automated system.[67] By performing direct susceptibility test from positive blood culture bottles, for example, direct disk diffusion (dDD) test, the TAT to generate antibiotic sensitivity report can reduce to 24 h. However, there is a grave need to address the problems associated with the methodology of performing and reporting dDD.
Since 1980s, there are lot of studies regarding standardization of direct susceptibility testing in various platforms such as disk diffusion, automated, and molecular techniques.[891011] Even with all these studies, there are no standard international documents regarding the dDD testing technique, due to which many clinical laboratories still hesitate to put dDD results on patients chart.[7] In 2014, Clinical and Laboratory Standards Institute (CLSI) Subcommittee on Antimicrobial Susceptibility Testing (AST) formed an ad-hoc working group to address the standardization of a direct-from-blood culture, and initial phase of study reports good categorical agreement (CA) of dDD with reference disk diffusion (rDD) test.[7] However, this was a simulated study performed on preserved isolates. More so, the CA between dDD and rDD may vary between laboratories. Therefore, this study was undertaken to determine the CA and understand the various type of errors associated with antibiotic class and pathogen combination, so that caution can be derived while interpreting and reporting dDD results in the earliest meaningful timeframe.
Materials and Methods
The study was conducted in a tertiary care public sector 2200 bedded hospital, South India from August 2017 to January 2018 (6 months). All the positive aerobic blood cultures from patients suspected of having bloodstream infections (BSIs) were subjected to Gram staining. The specimens excluded from study were blood cultures which grew more than one type of isolate and blood culture in which Gram stain smear revealed budding yeast cells. dDD was performed according to CLSI guideline.[12] Four drops of blood culture broth (from a venting needle) were inoculated onto the Mueller-Hinton agar (MHA) plate, and lawn culture was performed using a sterile swab. After 15–20 min, antibiotic disks as per the direct Grams interpretation were applied on to the MHA surface, using a sterile forceps. Plates are read and interpreted as per CLSI breakpoints, after 18 h incubation.[13] rDD was performed from the isolate grown on the subculture plates on the next day according to the CLSI guidelines. Zone sizes for quality control strains were evaluated using CLSI quality control ranges published in CLSI M100 ED27-2017.[13]
Study design and analysis
dDD results were compared to the rDD results from the positive blood culture bottles. CA was evaluated, using breakpoints mentioned in CLSI M100 ED27-2017, after excluding any antimicrobials for which the pathogen known to have intrinsic resistance.[13]
Gram-negative bacilli were tested for panel of 8 antibiotics such as amikacin 30 μg (AK), gentamicin 10 μg (G), ciprofloxacin 5 μg (CF), ceftriaxone 30 μg (CTR), ceftazidime 30 μg (CAZ), cefoperazone sulbactam 75/30 μg (CFS), piperacillin-tazobactam 100/10 μg (PIT), and meropenem 10 μg. All these were included for analysis if the pathogen was identified as a member of Enterobacteriaceae family or Acinetobacter species. For Pseudomonas species, CTR was excluded from the analysis. All other nonfermenters were excluded from the analysis as the antibiotic panel was different from that used in this study.
The antibiotic panel used for Gram-positive cocci in clusters (suspected Staphylococcus species) comprised of 6 disks such as penicillin 10 units (PEN), cefoxitin 30 μg (OX), erythromycin 15 μg, co-trimoxazole 1.25/23.75 μg, CF 5 μg, and linezolid 30 μg (LZ) and for Gram-positive cocci in pairs (suspected Enterococcus species) ampicillin 10 μg (AMP), high level G 120 μg (HLG), tetracycline 30 μg, and LZ 30 μg.
The performance of dDD as compared to rDD was expressed in terms of CA and categorical disagreement. The categorical disagreement was further characterized into minor error (mE), major error (ME), and very ME (VME) as depicted in Table 1. All collected data were entered into Microsoft excel sheet. The analysis of data was carried out using IBM SPSS Statistics for Windows, Version 19.0. (Armonk, NY: IBM Corp).
| CA | Categorical disagreement | ||||||
|---|---|---|---|---|---|---|---|
| mE | ME | VME | |||||
| rDD | S | I | R | R or S | I | S | R |
| dDD | S | I | R | I | R or S | R | S |
R=Resistant, S=Sensitive, I=Intermediate, mE=Minor error, ME=Major error, VME=Very ME, rDD=Reference disk diffusion, dDD=Direct disk diffusion, CA=Categorical agreement
Terminologies used for comparison of performance of dDD test with rDD tests.
Results
The workflow in each laboratory is unique and in a routine workup of positive blood culture bottles varies across laboratories and is evolving with new technologies. In laboratories with increased sample load and limited resources, performing dDD and comparing with rDD still remains as the suitable option for abbreviating time to AST report.
As shown in Figure 1, 17, 215 blood cultures were received during the study, of which 71.9% (12,388) cultures were sterile, 12.7% (2194) of cultures were contaminated, 0.2% (27) of the blood cultures were rejected due to inappropriate requisition form or mislabeling of the bottle. The total pathogens were isolated 15.2% (2606); of which the following 57.2% (1491) were excluded from the study: (i) 44.4% (1157) of cultures, the direct bottle grams did not match culture smear, (ii) 6.3% (163) of cultures, dDD did not grow or MHA plate was contaminated, and (iii) 6.6% (171) of the cultures, the dDD was not performed as the bottle smear showed budding yeast cells. The remainder pathogens, 42.8% (1115) were considered for dDD and rDD tests.
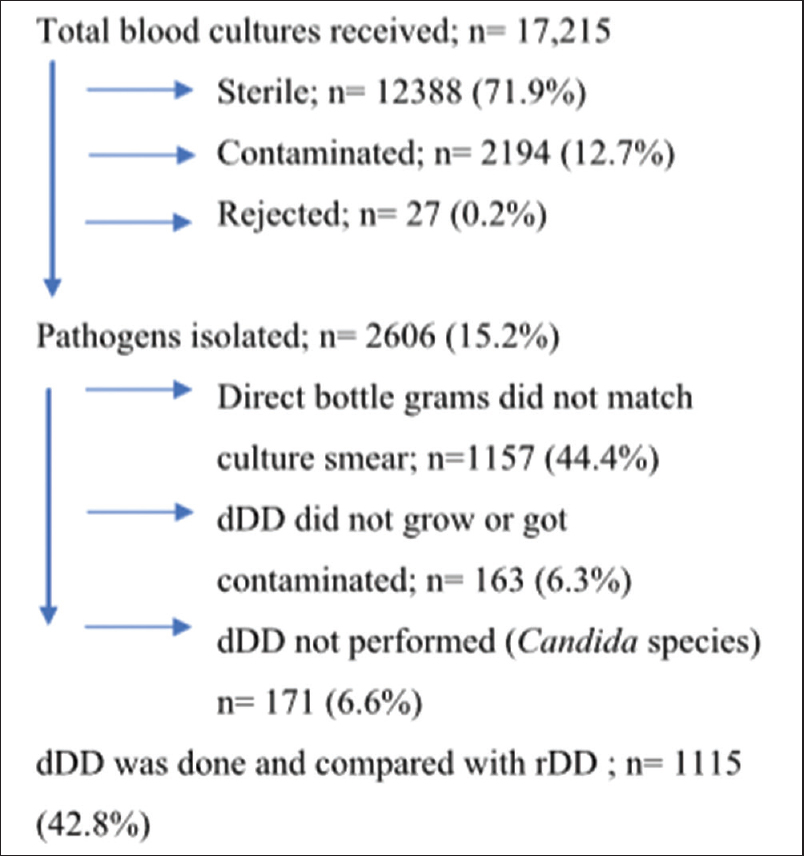
- Selection of isolates (pathogens) for performing direct disk diffusion (dDD) test and reference disk diffusion (rDD) test
Table 2 shows the distribution of bacteria isolated from the positive blood cultures for which both dDD and rDD tests were performed. Enterobacteriaceae accounts for 42.7% (476) of total isolates; nonfermenters 40% (446) and Gram-positive cocci 17.3% (193). Among Gram-negative bacilli, Escherichia coli was the most common isolate (18.9%), followed by Pseudomonas species (17.3%), Acinetobacter species (13.2%), and Klebsiella species (13%). Among Gram-positive cocci, Staphylococcus aureus was the most common isolate (7.8%) followed by coagulase-negative staphylococci (CoNS, 5%) and Enterococcus species (4%). For further analysis of CA between dDD and rDD, we have excluded the isolates which were <30 (Proteae tribe, Salmonella species, Serratia species, Pantoea species, and Streptococcus species) as analysis will not be significant and also non-fermenting Gram-negative bacilli other than Pseudomonas species and Acinetobacter species (other NF-GNB) as they were not further characterized and had an antibiotic panel different from routine Gram-negative antibiotic panel used in this study. Hence, the isolates included for analysis were 965.
| Organisms | Number of isolates tested, n (%) |
|---|---|
| Enterobacteriaceae | 476 (42.7) |
| Escherichia coli | 211 (18.9) |
| Klebsiella species | 145 (13) |
| Enterobacter species | 81 (7.3) |
| Proteae tribe* | 14 (1.3) |
| Salmonella species* | 12 (1.1) |
| Serratia species* | 11 (1) |
| Pantoea species* | 2 (0.2) |
| Nonfermenters | 446 (40) |
| Pseudomonas species | 193 (17.3) |
| Acinetobacter species | 147 (13.2) |
| Other NF-GNB** | 106 (9.5) |
| Gram-positive cocci | 193 (17.3) |
| Staphylococcus aureus | 87 (7.8) |
| CoNS | 56 (5) |
| Enterococcus species | 45 (4) |
| Streptococcus species* | 5 (0.4) |
| Total | 1115 |
Totally 965 isolates were analyzed after excluding the following organisms (i) Isolates* for which the numbers were<30; and (ii) Isolates** which were not further characterized and had a different antibiotic panel from routine Gram-negative antibiotic panel used in this study. NF-GNB=Nonfermenting Gram-negative bacilli, CoNS=Coagulase negative Staphylococcus
As shown in Table 3, overall, dDD performed excellent with a CA of 96% with rDD; mE of 0.9%, ME of 2.1% and VME of 1% fulfilling the performance criteria considered acceptable (ME ≤3%; VME ≤3%) by the international standard organization (ISO 20776-2).[714] VME was highest in Pseudomonas species (3%); ME and mE were highest in Enterobacteriaceae (2.8% and 1.2%, respectively). When analyzed for the disagreement at number of antibiotics per isolate; it was observed that most of the organisms had a disagreement at ≤2 antibiotics per isolate (10.7%); significant disagreement at ≥2 antibiotics was observed with Pseudomonas species (7.3%).
| Organisms and antibiotic tested (n×Ab=N) | CA, n (%) | Categorical disagreement, n (%) | |||||
|---|---|---|---|---|---|---|---|
| Among isolate-antibiotic combinations tested | Among the isolates tested | ||||||
| Minor | Major | Very major | Total | Disagreement at ≤2 antibiotics | Disagreement at ≥2 antibiotics | ||
| Enterobacteriaceae (437×8 = 3496) | 3342 (95.6) | 42 (1.2) | 98 (2.8) | 14 (0.4) | 154 (4.4) | 57 (13) | 23 (5.3) |
| Pseudomonas spp. (193×7 = 1351) | 1278 (94.6) | 13 (1) | 19 (1.4) | 41 (3) | 73 (5.4) | 19 (9.8) | 14 (7.3) |
| Acinetobacter spp. (147×8 = 1176) | 1135 (96.5) | 11 (0.9) | 17 (1.4) | 13 (1.1) | 41 (3.5) | 17 (11.6) | 6 (4.1) |
| Staphylococcus spp. (143×6 = 858) | 845 (98.5) | 0 | 12 (1.4) | 1 (0.1) | 13 (1.5) | 7 (4.9) | 1 (0.7) |
| Enterococcus spp. (45×5 = 225) | 222 (98.7) | 0 | 3 (1.3) | 0 | 3 (1.3) | 3 (6.7) | 0 |
| Overall (7106) | 6822 (96.0) | 66 (0.9) | 149 (2.1) | 69 (1.0) | 284 (4.0) | 103 (70.1) | 44 (29.9) |
n=Number of isolates, Ab=Number of antibiotics tested, N=Number of isolate-antibiotic combinations. CA=Categorical agreement
The analysis of errors of dDD compared to rDD is vital for carrying out selective reporting of dDD results. We studied this among Enterobacteriaceae, Pseudomonas species, Acinetobacter species, and Gram-positive cocci.
Among Enterobacteriaceae [Table 4], CA was >95% for all the antibiotics in the panel except for CFS (91.8%) and PIT (92.4%). VME among Enterobacteriaceae was well within the acceptable limits (VME ≤ 3%). Higher ME was observed for G (3.2%), CFS (5.5%), and PIT (5.5%).
| Enterobacteriaceae (n=437) | CA, n (%) | Categorical disagreement, n (%) | |||
|---|---|---|---|---|---|
| Minor | Major | Very major | Total | ||
| AK | 427 (97.7) | 0 | 9 (2.1) | 1 (0.2) | 10 (2.3) |
| G | 419 (95.9) | 0 | 14 (3.2) | 4 (0.9) | 18 (4.1) |
| CF | 430 (98.4) | 2 (0.5) | 4 (0.9) | 1 (0.2) | 7 (1.6) |
| CTR | 426 (97.5) | 2 (0.5) | 5 (1.1) | 4 (0.9) | 11 (2.5) |
| CAZ | 419 (95.9) | 9 (2.1) | 7 (1.6) | 2 (0.5) | 18 (4.1) |
| CFS | 401 (91.8) | 11 (2.5) | 24 (5.5) | 1 (0.2) | 36 (8.2) |
| PIT | 404 (92.4) | 9 (2.1) | 24 (5.5) | 0 | 33 (7.6) |
| MRP | 416 (95.2) | 9 (2.1) | 11 (2.5) | 1 (0.2) | 21 (4.8) |
CA=Categorical agreement, AK=Amikacin, G=Gentamicin, CF=Ciprofloxacin, CTR=Ceftriaxone, CAZ=Ceftazidime, CFS=Cefoperazone sulbactam, PIT=Piperacillin-tazobactam, MRP=Meropenem
Table 5 depicts that there was a CA of > 95% in Pseudomonas species for all the antibiotics tested except for AK (91.2%), G (92.2%) and CF (91.2%). VME was found to be marginally above the acceptable limits for AK (6.2%), G (5.2%) and CF (4.1%). ME among all antibiotics was within the justifiable limits. mE was high for CF (4.7%), but there are no cutoff limits for interpretation of mEs.
| Pseudomonas species (n=193) | CA, n (%) | Categorical disagreement, n (%) | |||
|---|---|---|---|---|---|
| Minor | Major | Very major | Total | ||
| AK | 176 (91.2) | 1 (0.5) | 4 (2.1) | 12 (6.2) | 17 (8.8) |
| G | 178 (92.2) | 0 | 5 (2.6) | 10 (5.2) | 15 (7.8) |
| CF | 176 (91.2) | 9 (4.7) | 0 | 8 (4.1) | 17 (8.8) |
| CAZ | 185 (95.9) | 1 (0.5) | 2 (1) | 5 (2.6) | 8 (4.1) |
| CFS | 188 (97.4) | 0 | 2 (1) | 3 (1.6) | 5 (2.6) |
| PIT | 187 (96.9) | 1 (0.5) | 2 (1) | 3 (1.6) | 6 (3.1) |
| MRP | 188 (97.4) | 1 (0.5) | 4 (2.1) | 0 | 5 (2.6) |
CA=Categorical agreement, AK=Amikacin, G=Gentamicin, CF=Ciprofloxacin, CAZ=Ceftazidime, CFS=Cefoperazone sulbactam, PIT=Piperacillin-tazobactam, MRP=Meropenem
For Acinetobacter species [Table 6], the CA was > 95% for all the antibiotics in the panel except for CFS (93.2%). VME and ME among Acinetobacter species were well within the acceptable limits; however, the mE was higher for CFS (3.4%).
| Acinetobacter species (n=147) | CA, n (%) | Categorical disagreement, n (%) | |||
|---|---|---|---|---|---|
| Minor | Major | Very major | Total | ||
| AK | 143 (97.3) | 1 (0.7) | 0 | 3 (2) | 4 (2.7) |
| G | 140 (95.2) | 1 (0.7) | 2 (1.4) | 4 (2.7) | 7 (4.8) |
| CF | 141 (96) | 3 (2) | 2 (1.4) | 1 (0.7) | 6 (4.1) |
| CTR | 144 (98) | 0 | 2 (1.4) | 1 (0.7) | 3 (2) |
| CAZ | 144 (98) | 0 | 1 (0.7) | 2 (1.4) | 3 (2) |
| CFS | 137 (93.2) | 5 (3.4) | 4 (2.7) | 1 (0.7) | 10 (6.8) |
| PIT | 142 (96.6) | 1 (0.7) | 3 (2) | 1 (0.7) | 5 (3.4) |
| MRP | 144 (98) | 0 | 3 (2) | 0 | 3 (2) |
CA=Categorical agreement, AK=Amikacin, G=Gentamicin, CF=Ciprofloxacin, CAZ=Ceftazidime, CFS=Cefoperazone sulbactam, PIT=Piperacillin-tazobactam, MRP=Meropenem
Among Gram-positive cocci, both Staphylococcus species [Table 7] and Enterococcus species [Table 8] had CA of > 95% for all the antibiotics tested. VME was unsatisfactory in Staphylococcus species for cefoxitin (4.9%) and for HLG (4.4%) in Enterococcus species. ME and mE were satisfactory among both groups.
| Staphylococcus species (n=143) | CA, n (%) | Categorical disagreement, n (%) | |||
|---|---|---|---|---|---|
| Minor | Major | Very major | Total | ||
| PEN | 141 (98.6) | 0 | 1 (0.7) | 1 (0.7) | 2 (1.4) |
| OX | 137 (95.8) | 0 | 7 (4.9) | 0 | 7 (4.9) |
| CF | 142 (99.3) | 0 | 1 (0.7) | 0 | 1 (0.7) |
| ERY | 140 (97.9) | 0 | 3 (2.1) | 0 | 3 (2.1) |
| LZ | 143 (100) | 0 | 0 | 0 | 0 |
CA=Categorical agreement, CF=Ciprofloxacin, LZ=Linezolid, PEN=Penicillin, OX=Cefoxitin, ERY=Erythromycin
| Enterococcus species (n=45) | CA, n (%) | Categorical disagreement, n (%) | |||
|---|---|---|---|---|---|
| Minor | Major | Very major | Total | ||
| AMP | 44 (97.8) | 0 | 1 (2.2) | 0 | 1 (2.2) |
| HLG | 43 (95.6) | 0 | 2 (4.4) | 0 | 2 (4.4) |
| TET | 45 (100) | 0 | 0 | 0 | 0 |
| Vancomycin | 45 (100) | 0 | 0 | 0 | 0 |
| LZ | 45 (100) | 0 | 0 | 0 | 0 |
CA=Categorical agreement, LZ=Linezolid, AMP=Ampicillin, G=Gentamicin, HLG=High level G, TET=Tetracycline
Discussion
Rapid identification and antimicrobial susceptibility results of organisms causing BSIs are an absolute priority for the microbiology laboratory. By performing dDD from positive blood cultures, clinicians can get adequate information to tailor the empirical treatment towards targeted antibiotic therapy about 24 h earlier than the conventional rDD test. This in turn can lead to a substantial reduction in mortality and morbidity of the patient.[15]
CLSI, British Society of Antimicrobial Chemotherapy Guidelines, and European Committee on AST have proposed several methods to perform dDD.[6716] However, there is a lack of large scale studies and guidelines on reporting of dDD results. Furthermore, the CA and disagreement can have interlaboratory variation based on the methodology followed for performing dDD and the type of automated blood culture bottles used. We designed the present study to determine the potential accuracy of dDD testing.
In the present study, we evaluated 965 pathogens and 7106 organism-antibiotic combinations. Overall, there was a CA of 96% which was extremely satisfactory. The categorical disagreement was found only in 4% of organism-antibiotic combinations; the majority of which were ME (2.1%) followed by VME (1%) and mE (0.9%). Reporting false resistance (in case of ME) will not put the patient into any stake, but may promote drug resistance which can be obviated by verbal communication of rDD results immediately. Percentages of errors (mE, ME, and VME) were overall much lower than the acceptable performance criteria of International Standard ISO 20776-2 (ME ≤3%; VME ≤3%).[714] A study on Gram-negative bacteria conducted by Chandrasekaran et al. in the initial results of CLSI study group showed a CA of 87.9% between dDD and rDD; which was lower to that observed in our study. The errors found in this study were VME (0.5%), ME (3.5%), and mE (10%). The present study showed a better performance of dDD with rDD which could be attributed to the large number of diverse clinical isolates tested in our study including Gram-positive organisms. Desai et al. reported the overall CA of 90.4% between dDD and rDD with 1.8% VME, 1.9% ME, and 5.8% mE among Gram-negative bacteria.[17]
Except the two studies quoted above, there is paucity of recent literature comparing dDD with rDD, as most of the studies focused on the comparison of dDD with AST from colonies by automated systems (Vitek, Phoenix, or Micro scan). In a study conducted in an oncology center from Kolkata, Goel et al. reported CA of 83.7% between dDD compared with AST from colonies (Gram-negative) by Vitek-2.[9] In another multicenter study, Coyle et al. reported CA of 94.3% for dDD tested with Gram-positive bacteria.[18] As a novel initiative, we analyzed the categorical disagreement at ≤2 and ≥2 antibiotics. We observed that the categorical disagreement at ≤2 and ≥2 antibiotics were 70.1% and 29.9%, respectively, by which we can derive that even if there is categorical disagreement, majority will have discrepancies with ≤2 antibiotics.
When the performance of dDD with rDD in Enterobacteriaceae was evaluated, we found very good results with CA above 90%. CA was least with beta-lactamase inhibitor group (BLI), i.e., with CFS (91.8%) and PIT (92.4%) with ME of 5.5% in both. Similar findings were also observed by Chandrasekaran et al.[7] in a CLSI working group study; reported a CA of 83.3% for PIT and Desai et al.[17] who reported CA of 71.7% for BL-BLI (ampicillin-sulbactam).[17] Unlike our study, in both these studies, the disagreement was attributed to mE. On assessing the performance of dDD with rDD in Pseudomonas species, we observed CA above 90% for all organism-antimicrobial combinations. VME was reported above the acceptable range in AK (6.2%), G (5.2%), and CF (4.1%). In discordance to our results, Goel et al. reported a low CA for CAZ (76.1%) and this variation was attributed to VME.[9] For Acinetobacter species CA was extremely satisfactory without any significant errors.
For Staphylococcus species, the findings were satisfactory except for cefoxitin (CA 95.8%, ME 4.9%). A similar observation was seen in a study conducted by Bennet et al.,[19] where they reported an agreement of 88% for cefoxitin dDD with rDD. We recommend to report cefoxitin dDD for methicillin-resistant S. aureus (MRSA) and MR-CoNS, however caution should be taken, and clinicians can be communicated if any discrepancies. More so, a supplemental test like MRSA latex can be performed to confirm the result without affecting the TAT. HLG for Enterococcus species had an ME of 4.4%. To the best of our knowledge, no other literature was available to compare the results.
The initial results of CLSI study group also showed that variables such as blood culture incubation monitoring systems (BacT/ALERT, BACTEC, and Versa TREK systems) and concentration of bacteria in the broth inoculum are the major discrepancies hindering the performance of dDD. In our study, we did not evaluate these variables, which could be our future venture.[7]
Conclusion
Routine blood culture practice is optimal if bottles are promptly placed on blood culture instrument (within 2 h of collection), punctually removed soon after it flags and the direct Gram stain and antimicrobial susceptibility results communicated rapidly to the clinicians so that they respond expeditiously. dDD is of paramount importance for early institution of targeted therapy and is considered as one of the key stewardship intervention. Our study in concordance to many other studies including the initial report of CLSI working group 2018 give an insight that every laboratory must perform dDD for positively flagged blood culture specimens; the result of which should be confirmed later by performing rDD. We also conclude that one should be vigilant while reporting dDD result of BL-BLI for Enterobacteriaceae; aminoglycosides and CF for Pseudomonas species; cefoxitin for Staphylococcus species and HLG for Enterococcus species. Supplementary tests such as MRSA latex should be included when necessary.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43:304-77.
- [Google Scholar]
- Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589-96.
- [Google Scholar]
- First notification of positive blood cultures and the high accuracy of the gram stain report. J Clin Microbiol. 2007;45:1113-7.
- [Google Scholar]
- 2014. UK Standards for Microbiology Investigations Investigation of Blood Cultures (for Organisms other than Mycobacterium species) [Internet]. Available from: http://www.apsi.it/public/ufiles/smi/b37_8_en_141104.pdf
- Update on blood cultures: How to obtain, process, report, and interpret. Clin Microbiol Infect. 2013;19:513-20.
- [Google Scholar]
- Direct disk diffusion test using European clinical antimicrobial susceptibility testing breakpoints provides reliable results compared with the standard method. Eur J Microbiol Immunol (Bp). 2015;5:103-11.
- [Google Scholar]
- Direct-from-blood-culture disk diffusion to determine antimicrobial susceptibility of Gram-negative bacteria: Preliminary report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol. 2018;56 pii: e01678-1
- [Google Scholar]
- Evaluation of a direct blood culture disk diffusion antimicrobial susceptibility test. Antimicrob Agents Chemother. 1981;20:696-8.
- [Google Scholar]
- A method for early detection of antibiotic resistance in positive blood cultures: Experience from an oncology centre in eastern India. Indian J Med Microbiol. 2015;33:53.
- [Google Scholar]
- Direct susceptibility testing with positive BacT/Alert blood cultures by using MicroScan overnight and rapid panels. J Clin Microbiol. 1998;36:2052-6.
- [Google Scholar]
- Comparison of Antimicrobial Susceptibility Testing of Isolates from Blood Cultures by Direct Inoculation Method and PHOENIX. J Biomed Lab Sci. 2011;23:23-7.
- [Google Scholar]
- DRAFT: Direct Susceptibility Testing of Gram Negative Rods from Blood Cultures [Internet]. Available from: www.jscm.org/kokusai/document/2016clsi_2.pdf
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI Document M100 (27th ed). Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- [Google Scholar]
- 2009. Guidance for Industry and FDA Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems [Internet]. Available from: https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm071462.pdf
- The clinical and prognostic importance of positive blood cultures in adults. Am J Med. 2010;123:819-28.
- [Google Scholar]
- Can direct disk diffusion susceptibility testing from positive blood cultures provide earlier results to clinicians? Open Forum Infect Dis. 2016;3(Suppl 1):180.
- [Google Scholar]
- Rapid antimicrobial susceptibility testing of isolates from blood cultures by direct inoculation and early reading of disk diffusion tests. J Clin Microbiol. 1984;20:473-7.
- [Google Scholar]
- Rapid differentiation of methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus from blood cultures by use of a direct cefoxitin disk diffusion test. J Clin Microbiol. 2008;46:3836-8.
- [Google Scholar]





