Translate this page into:
Impact of nucleic acid testing in double screening of blood donations in Northern India: A single-center experience
*Corresponding author: Sonia Gupta, Department of Immunohaematology and Blood Transfusion, Dayanand Medical College and Hospital, Ludhiana, Punjab, India. sonia4334@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Kumar R, Gupta S, Kaur H, Kaur S, John JM. Impact of nucleic acid testing in double screening of blood donations in Northern India: A single-center experience. J Lab Physicians. 2024;16:338-46. doi: 10.25259/JLP_4_2024
Abstract
Objectives:
The aim of our study is to assess the ability of nucleic acid testing (NAT) to detect hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus (HIV) from donor blood samples which were declared sero-non-reactive.
Materials and Methods:
The whole blood donations were collected over the 6-year period from 2017 to 2023 which were initially screened for hepatitis B surface antigen, anti-HCV, anti-HIV-1/2, and p24 antigen and syphilis by electrochemiluminescence assays by Roche Diagnostics, Germany and malaria by Pan/Pf rapid test. Donations were declared nonreactive by these tests which were further subjected to NAT testing using minipool method. It was done in pool of 6 using cobas Taq Screen MPX version 2.0 on the cobas s201 platform. The NAT yields were quantitated for viral loads and followed up by serology.
Statistical analysis:
The data obtained was entered in Microsoft Excel and analyzed using descriptive statistics using the Statistical Package for the Social Science 21 version.
Results:
Out of 152,575 donations, 149,304 were sero-non-reactive and screened by NAT. Out of 82, 45 were reactive for HBV deoxyribonucleic acid and 37 for HCV ribonucleic acid. The NAT yield was 1:1831 overall, 1:3337 for HBV and 1:4059 for HCV. Viral load was quantitated in 65/82 NAT yields (35 HBV and 30 HCV). The viral loads of HBV samples were <20 IU/mL for 19/35 samples and the HCV sample viral loads ranged from 25.3 to 9.3 × 106 IU/mL. Eleven NAT-yield donors (4 HBV, 7 HCV) reported for follow-up showed sero-conversion between 90 and 210 days after NAT screening.
Conclusions:
This study confirmed the benefit of a highly sensitive minipool NAT (MP-NAT) in interdicting infectious donations undetected by conventional screening and illustrates the detection of donations with low viral loads using the MP-NAT polymerase chain reaction.
Keywords
Blood safety
Transfusion-transmitted infections
Nucleic acid testing
Minipool-nucleic acid testing
INTRODUCTION
India has an annual requirement of 12 million units for transfusion.[1] Due to the high prevalence of infections in the donor and general population in India, especially with respect to the three major transfusion-transmitted infections (TTIs) of viral origin, namely, hepatitis B virus (HBV) hepatitis C virus (HCV) and human immunodeficiency virus (HIV),[2] serological screening is mandatory for the three viral TTIs in blood banks in India.[3] Even though the use of advanced serological techniques improves the sensitivity of detection of TTIs, complete reliance on serology-only screening techniques still leads to missing of some of the donations harboring infections.[4,5] The conventional serological screening assays have wide “window period” of detection and may also miss occult hepatitis B infections (OBI), immune-silent carriers, and immunologically variant viruses.[6] Consequently, the Drugs and Cosmetics Act and the National AIDS Control Organization (NACO) allow for the introduction of better techniques to improve blood safety.[7,8]
The national seroprevalence of hepatitis B and hepatitis C is 0.95% (0.89–1.01) and 0.32% (0.28–0.36), respectively.[9] Moreover, the distribution of the disease burden among the states is unequal. The seroprevalence of HCV in Punjab has been found to be about 3.29% in the general population.[10] Punjab, at 1.35%, also has the highest seropositivity for hepatitis C among blood donors, much higher than the national average of 0.34%. In fact, unlike the rest of the country, the seropositivity of hepatitis C is higher than that of hepatitis B (0.65%) in blood donors in Punjab.[11,12] The heightened prevalence of HCV in Punjab can be traced to a combination of factors including insecure medical practices, unsafe therapeutic injections and surgeries, intravenous drug use, blood transfusions, hazardous dental procedures, and risky sexual practices.[13]
Molecular screening of HBV, HCV, and HIV by nucleic acid testing (NAT), in addition to serology, can improve the sensitivity of screening. It offers the potential of reduction of transmission of infections through transfusion.[14] A donated blood sample in which serological assay failed to detect the infection, but NAT detects a viral genome is referred to as a NAT yield.[15,16] The use of automated systems offers additional benefits of simplification of requirements of facilities in the blood center. The introduction of technological advances and automated molecular screening systems requires an investigation of their potential benefit.
This study reports the 6 year experience of implementing minipool NAT (MP-NAT) in a tertiary care hospital in North India. The data include the incremental yield obtained by employing a highly sensitive NAT in minipool format to serononreactive blood donations and subsequent quantitation of viral targets.
MATERIALS AND METHODS
Study setting
Blood donations were received in Department of ImmunoHematology and Blood Transfusion, Dayanand Medical College and Hospital, Ludhiana, Punjab, India, from April 2017 to March 2023 were included in the study. All blood donations were taken after routine consent as per NACO guidelines.
Serological screening
All the collected donations underwent initial screening by routine serology. Serological screening was done for hepatitis B surface antigen (HBsAg) (Elecsys HBsAgII), anti-HCV (Elecsys Anti-HCV-II), anti-HIV-1, anti-HIV-2, and HIV-p24 antigen (Elecsys HIV-combi PT) and syphilis (Elecsys Syphilis) by electrochemiluminescence technology on cobas e411 system (Roche Diagnostics, Germany). Screening for malaria was done by Pan/Pf rapid test (Malascan Plus, Viola Diagnostic System, India). All assays were performed as per the manufacturers’ instructions.
Molecular screening
Only donations that showed non-reactivity to screening for all serology targets were further tested by NAT. NAT was performed using the cobas® TaqScreen MPX Test version 2.0 (MPX2; Roche Molecular Systems, Branchburg, NJ, USA) on the cobas s201 platform (Roche Instrument Center, Rotkreuz, Switzerland). The system includes an automated sample pooling on the Hamilton MICROLAB® STARlet IVD Pipettor, automated sample preparation on the cobas Ampli Prep (CAP) instrument, and automated amplification and detection using the cobas TaqMan (CTM) analyzer. The MPX2 is a multiplex, multidye test employing polymerase chain reaction (PCR), for the simultaneous detection and identification of HBV, HCV, and HIV through detection of the targets HBV deoxyribonucleic acid (DNA), HCV ribonucleic acid (RNA), HIV-1 group M, HIV-1 group O, and HIV-2RNA.
Screening was performed on minipools of 6 samples. Reactive minipools were resolved by individually testing all 6 units of the minipool to identify the reactive donations and the virus. NAT was performed following the manufacturer’s instructions for the assay on an automated platform. The screening algorithm is summarized in Figure 1. The 95% and 50% detection limits for MPX2 are shown in Table 1.[17-20]
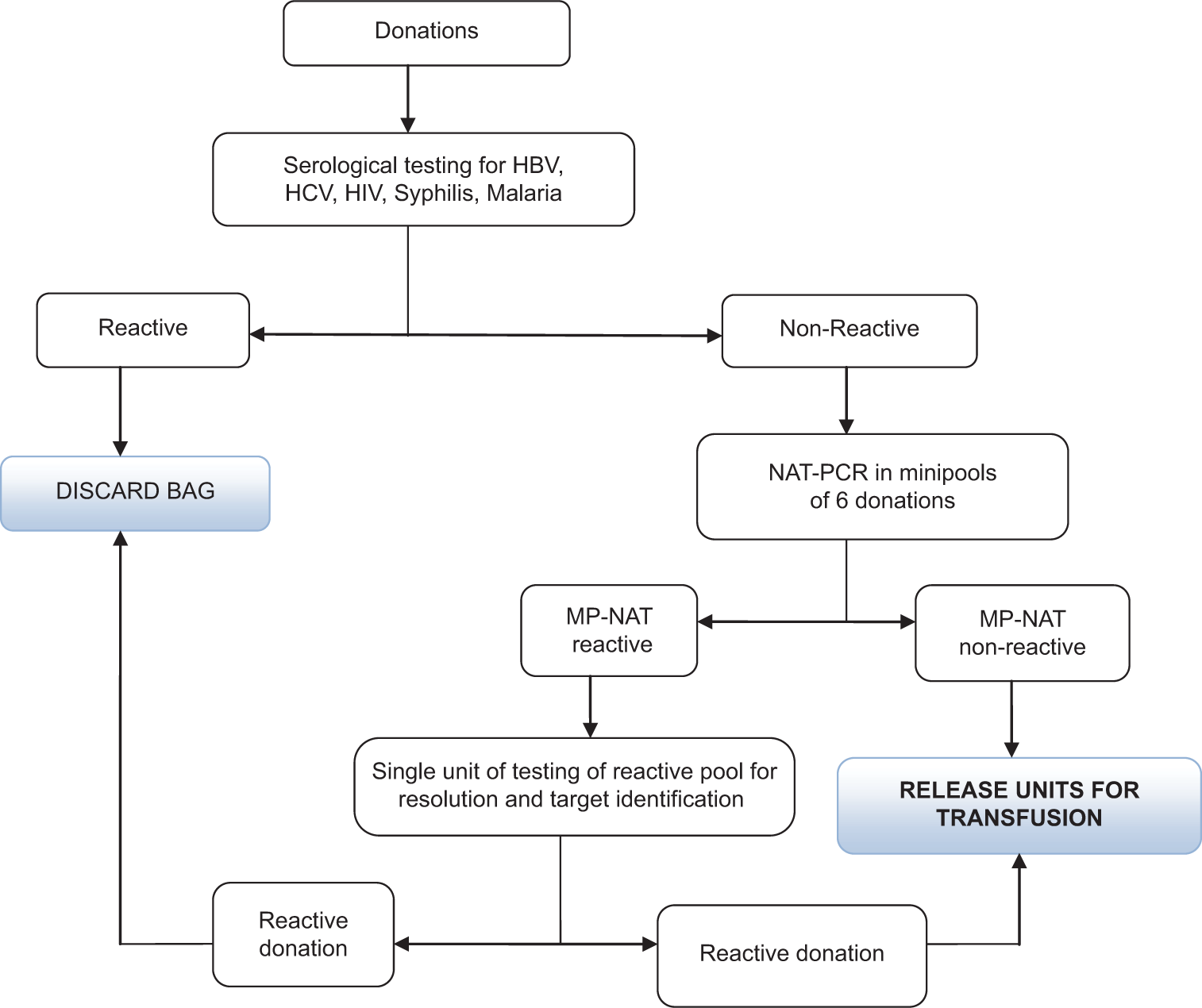
- Flow chart of minipool-nucleic acid testing algorithm. HBV: Hepatitis B, HCV: Hepatitis C, HIV: Human Immuno deficiency Virus, NAT-PCR: Nucleic Acid Testing-Polymerase chain reaction, MP- NAT: Minipool-Nucleic Acid Testing
| Target | cobas® TaqScreen MPX Test, v2.0donor screening assay | cobas® AmpliPrep/cobasTaqManviral load assays | ||
|---|---|---|---|---|
| 95% LOD*[16] | 50% LOD*[17] | LLOQ[18,19] | 95% LOD[18,19] | |
| HIV-1 Group M* | 50.3 IU/mL | 9.4 IU/mL | - | - |
| HCV | 6.8 IU/mL | 1.3 IU/mL | 15 IU/mL | 11 IU/mL |
| HBV | 2.3 IU/mL | 0.46 IU/mL | 20 IU/mL | 8.2 IU/mL |
LOD: Limit of detection, LLOQ: Lower limit of quantification, HIV: Human immunodeficiency virus, HBV: Hepatitis B virus, HCV: Hepatitis C virus. * LODs for HIV-1 Group O and HIV-2 are not shown here
Viral nucleic acid quantitation
The NAT yield donations were further evaluated for quantification of viral load using retained samples from frozen storage (−40°C). Samples were sent in the same order in which they were collected, as resources allowed. Samples were stored for a maximum of 3 months. Testing was performed using real time PCR by the CTM HBV test v2.0 and HCV test v2.0 run on a CTM 48 Analyzer (Roche Molecular Systems, Branchburg, USA). Table 1 shows the limits of detection and quantification for the viral load assays.
Follow-up testing of NAT yields
Donors of NAT-reactive donations were asked to return for follow-up serology at 90 days after NAT screening. Serological testing was performed with the same assays used for the initial serology screening on those reporting for follow-up.
Quality assurance
Our blood center is participating in External Quality Assessment Scheme, an external quality assurance program.
Statistical analysis
The data so obtained were entered into Microsoft Excel and analyzed using descriptive statistics. All statistical calculations were done using the Statistical Package for the Social Science 21 version statistical program for Microsoft Windows.
RESULTS
Serology
A total of 152,575 blood donations were screened by serology between April 2017 and March 2023. Of these, 4481 (2.93%) were seroreactive, as shown in Table 2. The remaining 148,094 donations were “serononreactive.” The highest seroreactivity was observed for HCV (2213/4481, 49.3%), followed by syphilis (1210/4481, 27.0%), HBV (867/4481, 19.3%), and HIV (191/4481, 4.26%).
| Screening target | Screen reactive by serology (%) |
|---|---|
| Total samples | 1,52,575 (100) |
| HBV surface antigen | 867 (0.56) |
| HCV antibody | 2213 (1.45) |
| HIV antigen/antibody | 191 (0.13) |
| Syphilis | 1210 (0.80) |
| Malaria | 0 (0) |
| Total | 4481 (2.93) |
HIV: Human immunodeficiency virus, HBV: Hepatitis B virus, HCV: Hepatitis C virus
Molecular screening
149,304 sero-non-reactive donations excluding syphilis were screened for viral nucleic acids, out of which 82 (0.05%) donations were found to be NAT reactive (NAT yields). The NAT yields by year are given in Table 3.
| Time period | NAT tested | Number of NAT-positive donations | NAT yield per donations tested | ||||
|---|---|---|---|---|---|---|---|
| HBV | HCV | Total | HBV | HCV | Total | ||
| April 2017–Mar 2018 | 25,023 | 11 | 12 | 23 | 1:2275 | 1:2085 | 1:1088 |
| April 2018–Mar 2019 | 24,316 | 7 | 10 | 17 | 1:3474 | 1:2432 | 1:1430 |
| April 2019–Mar 2020 | 25,780 | 4 | 5 | 9 | 1:6445 | 1:5156 | 1:2864 |
| April 2020–Mar 2021 | 22,003 | 10 | 2 | 12 | 1:2200 | 1:11001 | 1:1833 |
| April 2021–Mar 2022 | 24,861 | 8 | 5 | 13 | 1:3163 | 1:5061 | 1:1946 |
| April 2022–Mar 2023 | 27,321 | 5 | 3 | 8 | 1:1553 | 1:9256 | 1:3471 |
| Total | 149,304 | 45 | 37 | 82 | 1:3337 | 1:4059 | 1:1831 |
NAT: Nucleic acid testing, HBV: Hepatitis B virus, HCV: Hepatitis C virus
Viral load
Out of the total 82 NAT yield donations, we could evaluate viral loads for 65 donations. Of these, 35 were HBV reactive, and 30 were HCV reactive on NAT, as shown in Table 4. Results recorded as <20 IU/mL were reported as such by the assay, meaning that HBV DNA was detected but could not be quantified as the load was below the lower limit of quantitation of the assay.
| S. No. | NAT yield virus | Viral load (IU/mL) | S. No. | NAT yield virus | Viral load (IU/mL) |
|---|---|---|---|---|---|
| 1 | HBV | <20 | 36 | HCV | 25.3 |
| 2 | HBV | <20 | 37 | HCV | 1.25×103 |
| 3 | HBV | <20 | 38 | HCV | 7.49×103 |
| 4 | HBV | <20 | 39 | HCV | 1.05×104 |
| 5 | HBV | <20 | 40 | HCV | 3.49×104 |
| 6 | HBV | <20 | 41 | HCV | 7.03×104 |
| 7 | HBV | <20 | 42 | HCV | 7.51×104 |
| 8 | HBV | <20 | 43 | HCV | 9.01×104 |
| 9 | HBV | <20 | 44 | HCV | 1.01×105 |
| 10 | HBV | <20 | 45 | HCV | 3.02×105 |
| 11 | HBV | <20 | 46 | HCV | 4.82×105 |
| 12 | HBV | <20 | 47 | HCV | 4.93×105 |
| 13 | HBV | <20 | 48 | HCV | 5.72×105 |
| 14 | HBV | <20 | 49 | HCV | 5.72×105 |
| 15 | HBV | <20 | 50 | HCV | 2.2×106 |
| 16 | HBV | <20 | 51 | HCV | 2.29×106 |
| 17 | HBV | 1.07×103 | 52 | HCV | 2.59×106 |
| 18 | HBV | 1.5×103 | 53 | HCV | 4.69×106 |
| 19 | HBV | 6.3×104 | 54 | HCV | 4.7×106 |
| 20 | HBV | TND | 55 | HCV | 7.04×106 |
| 21 | HBV | <20 | 56 | HCV | 9.3×106 |
| 22 | HBV | 5.3×103 | 57 | HCV | TND |
| 23 | HBV | 6.4×104 | 58 | HCV | 1.03×103 |
| 24 | HBV | 7.3×103 | 59 | HCV | 6.2×103 |
| 25 | HBV | 4.02×103 | 60 | HCV | 5.03×103 |
| 26 | HBV | <20 | 61 | HCV | 7.2×104 |
| 27 | HBV | 7.06×103 | 62 | HCV | 6.2×104 |
| 28 | HBV | 9.03×103 | 63 | HCV | 3.2×103 |
| 29 | HBV | 8.5×104 | 64 | HCV | 4.0×103 |
| 30 | HBV | <20 | 65 | HCV | 8.0×104 |
| 31 | HBV | 1.09×103 | |||
| 32 | HBV | 2.2×103 | |||
| 33 | HBV | 5.5×103 | |||
| 34 | HBV | 2.2×103 | |||
| 35 | HBV | 5.6×103 |
NAT: Nucleic acid testing, HBV: Hepatitis B virus, HCV: Hepatitis C virus, TND: Target not detected
Follow-up testing
Eleven of the 82 NAT reactive donors reported for follow-up. The donors reported between 3 and 7 months (90–210 days) after NAT screening though they were called at 90 days. All 11 of these donors showed sero-conversion for the respective NAT reactive virus with follow-up ECLIA screening: four donors with HBV, and seven donors with HCV.
DISCUSSION
The data presented in this study demonstrate the ability of NAT applied in minipools to detect viral infections that are missed by serology. The data on viral load quantification and follow-up testing further indicate the potential high risk of transmission through the blood donations declared serononreactive, that were averted by NAT testing. Our study showed a high seroreactivity of 1.45% among all donors with HCV. Studies in the past have reported HCV seroprevalence among blood donors in Punjab to be around 1.38%,[21] and 1.45%.[22] As per the NACO/National Blood Transfusion Council (NBTC) report in 2016, the anti-HCV reactivity among donors in the blood center in Punjab was 1.35% in 2016.[11] This high seroreactivity for HCV in Punjab, where our blood center is located, is a reflection of the high seroprevalence of HCV (3.2%) in the general population of Punjab.[13] The higher seroreactivity observed for HCV in our study could be due to the use of a highly sensitive serological assay using the advanced ECLIA technology. Interestingly, the second highest seroreactivity in our study was observed for syphilis at 0.80%. Although blood center in Punjab has a high seroreactivity of 0.49% for syphilis,[11] the rate observed in our study is higher which could be again due to the use of a highly sensitive chemiluminescence based assay as against the rapid plasma reagin (RPR) test commonly employed in blood banks in the state. The seroreactivity for HBV and HIV was similar to other blood banks in the state.[11]
During the study, NAT in minipools was able to detect 45 HBV and 37 HCV infections from the donations which were “sero-non-reactive.” The NAT yields were thus 1:3337 for HBV, 1:4059 for HCV, and 1:1831 overall, as shown in Table 3.
The combined NAT yields reported from Indian tertiary care settings and stand-alone blood center are quite varied, and range between 1:476 and 1:4403 as per a 2017 review of 11 Indian studies.[23] A few studies have reported higher NAT yields of 1:354[24] and 1:974;[25] both the investigators used enzyme-linked immunosorbent assay (ELISA)-based serological testing. Chandra et al. also demonstrated a decrease in NAT yield by employing chemiluminescence on donations previously tested using ELISA.[24] Our study reported a higher overall NAT yield (1:1831) than most recent studies, and this could be due to differing prevalence of TTIs. Thus, a high NAT yield for HBV and a higher than usual HCV NAT yield have pushed the combined NAT yield higher in our study. Of note, the NAT yield in this study is comparable to or higher than other studies despite the fact that many other studies have employed individual donation NAT (ID-NAT), whereas we have used MP-NAT.
A high NAT reactivity was observed in our study over and above a high seroreactivity obtained with an advanced serological technology like ECLIA. This indicates a significant benefit of NAT in helping to minimize the risk of infectious donations entering the blood supply. Since each blood bag may be separated into multiple components for transfusion, each NAT yield has the potential to prevent transmission of TTIs to multiple transfusion recipients.
A noteworthy finding of our study is the result of the viral load quantitation in NAT yield samples. However, the initial plan was to quantify all NAT yields; viral load testing could only be done on 65/82 NAT yields due to inadequate sampling and clotting in a few samples. However, some interesting insights were revealed from this testing. The HCV NAT yields in our study showed very high viral loads ranging from 25.3 to 9.3 × 106. The high HCV viral loads observed in our study are consistent with the rapid doubling time of HCV in early infection. This demonstrates that donations with high viral titers of HCV, having a high potential for transmission, may be missed by serological assays detecting antibodies to HCV, further underlining the importance of narrowing the window period (WP) of detection through detection of the viral genome by NAT. In our study, the viral loads of 19/35 tested HBV NAT yields were below 20 IU/mL. These HBV infections with low viral loads that were missed by serology but picked up by NAT could be either OBI, recent HBV infections at a stage before the appearance of detectable levels of HBsAg, or resolving HBV infection at a stage where HBsAg has declined to below detectable levels but circulating HBV DNA is not yet cleared. There was one case each of HBV and HCV where NAT yield was positive, but the viral load was undetectable. One possible reason for this is that the sensitivities of the HBV and HCV quantitative molecular assays are lower than those for the MPX2 blood screening assay. In either of these cases, the viral load could possibly be below the limit of detection of the quantitative assays; however, the impact of sampling effects also cannot be completely ruled out.[26] The differences between the viral loads of HCV and HBV echo the differences in the biology of the two viruses.
Out of 82, 11 NAT reactive donors reported back after follow-up and were found to be positive for serology. According to NBTC guidelines,[27] all initial seroreactive and NAT positive donors should be recalled, given post donation counseling and repeat sample is to be taken for testing. 11 NAT initial reactive donors developed seropositivity (four donors with HBV and seven donors with HCV) and were counseled. The eventual appearance of HBsAg in the four HBV NAT reactive donors and anti-HCV in the seven HCV NAT reactive donors suggests that these were serological WP donations. In our study, all 11 donors with NAT-positive donations seroconverted, thereby affirming the value of NAT in averting certain TTI transmission. A similar finding was reported by Mishra et al. in 2017, where out of 51 seronegative samples showing NAT yield, seropositivity was observed within 6 months – 1 year for 38 donors (31/44 HBV, 2/2 HCV, and 5/5 HIV seroconversions).[28] In India, where the HBV endemicity is high, the frequency of OBI is often higher than acute infection. In 2014, Doda et al. evaluated the HBV antibody profiles of HBsAg negative, HBV DNA positive donations; out of 18 samples profiled, 12 showed antibodies against HBc (HBV core antigen), signifying OBI, whereas the remaining 6 had no detectable serological markers, indicating that these donors were in the WP of HBV infection.[29] The HCV NAT yields are majorly expected to be from the serological WP for HCV infection. While occult Hepatitis C infection is also described in the literature, the information about this entity is rather limited, and much research is needed in this area to understand the clinical implications of this condition.[30] A complete follow-up of all NAT yield donations would be a step toward a more comprehensive understanding of the stage of infection, the clinical implications of such donations, and mitigation of the risk.
Before the implementation of MP-NAT PCR in our set-up, we had been performing ID-NAT using the transcription-mediated amplification-based Procleix Ultrio Plus® (Novartis Diagnostic, USA) on the Procleix Panther system. The purpose of the current study was also to evaluate the overall ease of implementation of the fully automated NAT-PCR system. The cobas MPX2test is a multiplex PCR test that has the ability to simultaneously detect a reactive donation and identify the target virus (HBV, HCV, or HIV) with specific dyes in different channels. This eliminates the need for discriminatory testing in reactive donation samples for separately identifying the target virus and helps the blood bank plan further actions with certainty. Difficulty in confirming reactive donations obtained through IDNAT on the Ultrio Plus assay has been reported, including recently in India, where only 22%[31] and 41%[32] of initially reactive samples could be confirmed by repeat testing and viral target discrimination. In our previously published study of the ID-NAT assay, too, only about 83% of the initially reactive results were confirmed.[15] The algorithm that we used previously for the ID-NAT assay is given in Figure 2.[15] This discrepancy and non-confirmation lead to uncertainty on the cause of initial reactivity (non-specific amplification, sample contamination, low viral load, etc.) and the viral target, leading to subsequent difficulties in donor recall and counseling and unnecessary deferrals and repeat tests.[18] In comparison, we found the screening algorithm of the MPNAT PCR assay to be simpler. This algorithm uses NAT for testing only the sero-nonreactive donations; this reduces the number of tests to be carried out on reactive donor samples. Further, since the screening is done in minipools of multiple donations (in our case, six donations), there are additional benefits of fewer number of tests, higher specificity, and lower overall cost. Moreover, PCR is a time-tested technology. A few additional benefits that we found as a user of the MPNAT technology are minimal maintenance and hands-on time (since the system is completely automated), availability of ready-to-use reagents, no requirement of test calibrations, provision of internal control for monitoring test performance in each individual test, and incorporation of the AmpErase enzyme to reduce potential contamination by previously amplified material.
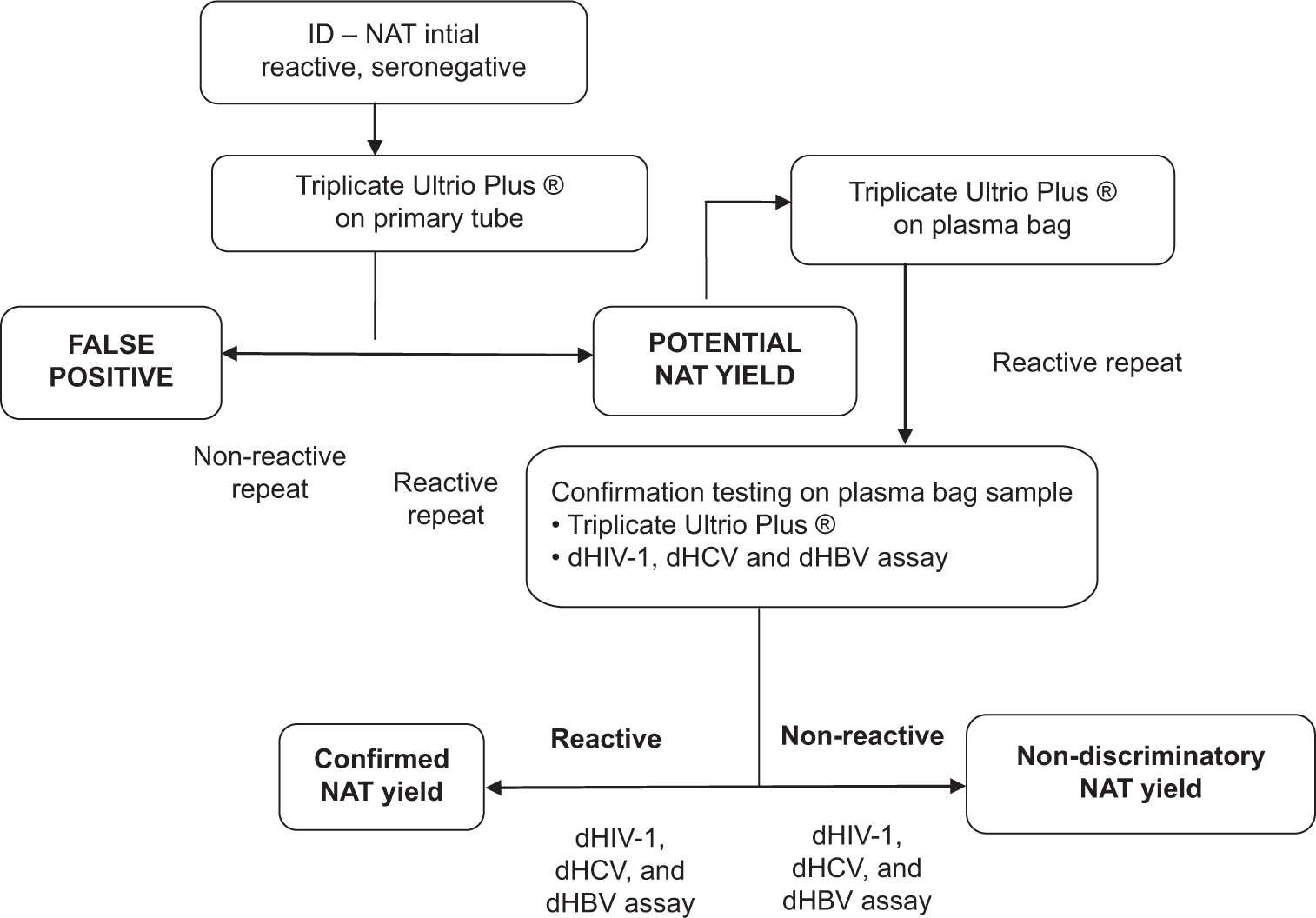
- Screening algorithm for individual donation-nucleic acid testing used by our blood bank previously. ID-NAT: Individual nucleic testing, dHIV: Discriminatory Human Immuno Deficiency Virus, dHCV: Discriminatory Hepatitis C, dHBV: Discriminatory Hepatitis B
The data generated by this study and their interpretation are reinforced by the size of the study, the additional quantitation data available, and follow-up serology. This large study demonstrated the utility of the highly sensitive cobas Taq Screen MPX2 to screen pooled samples and obtain high NAT yields. It also identified the viral loads and stage of infection of these donations, which can be used to derive the risk to blood safety and public and individual health. The investigation also illustrated the high convenience and ease of using automation and minipool testing. However, there are some limitations in the present study. The use of additional serological markers like antibody against Hepatitis C core antigen (HBc) in follow-up testing would have helped characterize OBIs. The use of a higher sensitivity quantitative molecular assay would have enabled us to demonstrate that the blood screening assay detects samples with much lower viral loads as has been shown by other authors.[24] Finally, the cost-effectiveness of the incorporation of MPNAT, in addition to serological testing and comparison to ID-NAT, was not assessed; further studies are required in this direction.
CONCLUSIONS
The present study has reconfirmed the high burden of TTIs in blood donations in India despite mandatory serological screening and has indicated that the routine application of MP-NAT screening in Indian blood centers would be a move one step closer to safer blood. Cardinal features required for adoption and smooth running in blood centers of varied sizes and donor infection prevalence include reliability, ensured by the use of a highly sensitive assay, ease of operation, ensured by the use of a fully automated system, and an uncomplicated testing algorithm. MP-NAT performed using the cobas MPX2 suitably fulfills both these conditions.
Ethical approval
The research/study was approved by the Institutional Ethics Committee, number IEC-2023-880 dated 10/07/2023.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- National AIDS Control Organization (NACO) 2014. Available from: http://www.naco.gov.in/sites/default/files/rapid%20situation%20assessment%20of%20bts%20in%20india%20pdf%20%281%29.pdf [Last accessed on 2024 Jan 08]
- [Google Scholar]
- Seroprevalence of transfusion transmitted infections in healthy blood donors: A 5-year tertiary care hospital experience. J Lab Physicians. 2017;9:283-7.
- [CrossRef] [PubMed] [Google Scholar]
- Seroprevalence of infectious markers and their trends in blood donors in a hospital based blood bank in north India. Indian J Med Res. 2015;142:317-22.
- [CrossRef] [PubMed] [Google Scholar]
- Transfusion transmitted infections in thalassaemics: Need for reappraisal of blood screening strategy in India. Transfus Med. 2014;24:79-88.
- [CrossRef] [PubMed] [Google Scholar]
- A comprehensive serological and supplemental evaluation of hepatitis B “seroyield” blood donors: A cross-sectional study from a tertiary healthcare center in India. Asian J Transfus Sci. 2015;9:189-94.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of occult hepatitis B and window period infection among blood donors by individual donation nucleic acid testing in a tertiary care center in South India. Pathog Glob Health. 2016;110:287-91.
- [CrossRef] [PubMed] [Google Scholar]
- National AIDS Control Organization. 2007. Available from: https://naco.gov.in/sites/default/files/standards%20for%20blood%20banks%20and%20blood%20transfusion%20services.pdf [Last accessed on 2024 Jan 08]
- [Google Scholar]
- Department of Health. 2016. Ministry of Health and Family Welfare, Govt of India. Available from: https://cdsco.gov.in/opencms/export/sites/cdsco_web/pdf-documents/acts_rules/2016drugsandcosmeticsact1940rules1945.pdf [Last accessed on 2024 Jan 08]
- [Google Scholar]
- Seroprevalence of hepatitis B and hepatitis C-//NVHCP. 2021. Available from: https://nvhcp.mohfw.gov.in>common_libs>ap [Last accessed on 2024 Jan 08]
- [Google Scholar]
- Towards a comprehensive national hepatitis prevention and control programme New Delhi, India: Country Office for India WHO; 2015.
- [Google Scholar]
- A report on the assessment of blood banks in Punjab, India. 2016. National Blood Transfusion Council. Available from: http://nbtc.naco.gov.in/assets/resources/reports/commonresource_1517228564.pdf [Last accessed on 2024 Jan 08]
- [Google Scholar]
- Assessment of NACO supported blood banks: A preliminary report 2016. 2016. National AIDS Control Organization (NACO). National Blood Transfusion Council (NBTC), New Delhi. Available from: http://www.naco.gov.in/sites/default/files/assessment%20of%20naco%20supported%20blood%20banks%20-%20a%20preliminary%20report%202016.pdf [Last accessed on 2024 Jan 08]
- [Google Scholar]
- Tackling the hepatitis C disease burden in Punjab, India. J Clin Exp Hepatol. 2016;6:224-32.
- [CrossRef] [PubMed] [Google Scholar]
- Nucleic Acid Technology (NAT) testing for blood screening: impact of individual donation and Mini Pool-NAT testing on analytical sensitivity, screening sensitivity and clinical sensitivity. ISBT Sci Ser. 2014;9:315-24.
- [CrossRef] [Google Scholar]
- Individual donor-nucleic acid testing for human immunodeficiency virus-1, hepatitis C virus and hepatitis B virus and its role in blood safety. Asian J Transfus Sci. 2015;9:199-202.
- [CrossRef] [PubMed] [Google Scholar]
- Confirmation and follow up of initial “NAT yields”: Prospective study from a tertiary healthcare center in India. Transfus Apher Sci. 2016;54:242-7.
- [CrossRef] [PubMed] [Google Scholar]
- Cobas® TaqScreen MPX Test version 2.0 for use on the cobas s 201 system Method Sheet 08092800001-02EN 2 Mannheim, Germany: Roche Diagnostics; 2019.
- [Google Scholar]
- Sensitivity and specificity of a new automated system for the detection of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus nucleic acid in blood and plasma donations. Transfusion. 2018;58:649-59.
- [CrossRef] [PubMed] [Google Scholar]
- Cobas® AmpliPrep/COBAS® TaqMan® HBV Test, version 2.0 for use with cobas AmpliPrep Instrument and cobas TaqMan analyzer or cobas TaqMan 48 method sheet 05382882001-08EN Branchburg, USA: Roche Molecular Systems, Inc.; 2018.
- [Google Scholar]
- Cobas® AmpliPrep/COBAS® TaqMan® HCV quantitative test, version 2.0 for use with cobas AmpliPrep instrument and cobas TaqMan analyzer or cobas TaqMan 48 method sheet 05902754001-07EN Branchburg, USA: Roche Molecular Systems, Inc.; 2016.
- [Google Scholar]
- Prevalence of markers of hepatitis C virus among the blood donors. J Clin Diagn Res. 2012;6:959-62.
- [Google Scholar]
- Prevalence of hepatitis C virus seropositivity among blood donors in a tertiary care hospital. Int Res J Med Sci. 2015;3:22-4.
- [Google Scholar]
- Nucleic acid amplification testing in Indian blood banks: A review with perspectives. Indian J Pathol Microbiol. 2017;60:313-8.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of HIV, HCV, HBV infection in blood donors detected by nucleic acid testing: An Indian experience. Eur J Pharm Med Res. 2016;3:278-92.
- [Google Scholar]
- Significance of nucleic acid testing in window period donations: Revisiting transfusion safety in high prevalence-low resource settings. J Pathol Nepal. 2016;6:906-9.
- [CrossRef] [Google Scholar]
- Detection and identification of occult HBV in blood donors in Taiwan using a commercial, multiplex, multi-dye nucleic acid amplification technology screening test. Vox Sanguinis. 2014;106:103-10.
- [CrossRef] [PubMed] [Google Scholar]
- Guidelines for blood donor selection and blood donor deferral - National Blood Transfusion Council (NBTC) 2017. Available from: https://naco.gov.in>sites>default>files>letter [Last accessed on 2024 Jan 08]
- [Google Scholar]
- NAT positivity in seronegative voluntary blood donors from western India. Transfus Apher Sci. 2017;56:175-8.
- [CrossRef] [PubMed] [Google Scholar]
- Serological characterization of occult hepatitis B virus infection among blood donors in India. Transfus Apher Sci. 2014;51:162-7.
- [CrossRef] [PubMed] [Google Scholar]
- Understanding occult hepatitis C infection. Transfusion. 2020;60:2144-52.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of two algorithms to confirm and discriminate samples initially reactive for nucleic acid amplification tests. Asian J Transfus Sci. 2017;11:140-6.
- [CrossRef] [PubMed] [Google Scholar]
- Initial trends of individual donation nucleic acid testing in voluntary and replacement donors from a tertiary care centre in north India. Indian J Med Res. 2019;149:633-40.
- [CrossRef] [PubMed] [Google Scholar]






