Translate this page into:
Molecular characterization and antimicrobial susceptibility profile of New Delhi metallo-beta-lactamase-1-producing Escherichia coli among hospitalized patients
Address for correspondence: Dr. Anjali Agarwal, Department of Microbiology, Hind Institute of Medical Sciences, Barabanki, Uttar Pradesh, India. E-mail: anjalikgmu@gmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Carbapenemase-producing microorganisms are becoming a major concern among hospital-acquired infections. There is also increased multidrug resistance seen among these isolates.
AIMS:
We have conducted this study to determine the prevalence of New Delhi metallo-beta-lactamase-1 (NDM-1) gene-producing Escherichia coli among hospitalized patients in a tertiary care hospital in Northern India.
SETTINGS AND DESIGN:
The study was conducted in the Department of Microbiology with the tertiary care hospital settings. It was a prospective cross-sectional observational study conducted during January 2014–August 2014.
MATERIALS AND METHODS:
A total of 500 nonduplicate E. coli samples were processed. The isolates with reduced susceptibility to ertapenem, i.e., zone diameter between 19 and 21 mm, were considered carbapenemase producers. These isolates were subjected to modified Hodge test for phenotypic confirmation. Polymerase chain reaction was performed on all the screened isolates for molecular detection of NDM-1 gene.
STATISTICAL ANALYSIS USED:
Chi-square test was used to analyze the data and P < 0.05 was considered statistically significant.
RESULTS:
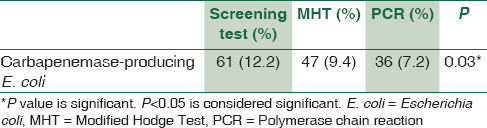
Out of 500 E. coli isolates, 61 (12.2%) were screened for carbapenemase production. 47 (9.4%) isolates were positive by modified Hodge test and 36 (7.2%) isolates showed the presence of blaNDM-1 gene (P < 0.05).
CONCLUSION:
There is an increased prevalence of NDM-1 gene-producing E. coli isolates. These carbapenemase-producing isolates are more resistant to other group of antibiotics (aminoglycosides, fluoroquinolones along with β-lactam group). Early detection of blaNDM-1 gene can help in choosing the effective treatment options for hospitalized patients in time, thereby reducing the risk of mortality.
Keywords
blaNDM-1 gene
carbapenemase
modified Hodge test
Introduction
Escherichia coli is one of the most common causes of hospital-acquired infections. E. coli can cause urinary tract infections, diarrhea, bloodstream infections, and pulmonary illnesses.[123] Till recently, the infections caused by these organisms could be treated effectively with the empirical antimicrobial therapies. Among these, third-generation cephalosporins were quite effective in the treatment of these infections with rare inclusion of carbapenems. However, due to unprecedented and irrational use of drugs, it has become more resistant to cephalosporins along with the carbapenems.[45678]
Carbapenemases are the enzymes included in group 2f (serine carbapenemases), 3a, and 3b (metallo-beta-lactamases) of Bush and Jacoby updated functional classification of bacterial beta-lactamases. Metallo-beta-lactamases (MBLs) differ from serine carbapenemases by their requirement of zinc ion at the active site. They have poor hydrolyzing capability for aztreonam. MBLs are not inhibited by clavulanate and tazobactam but inhibited by metal ion chelators such as ethylenediaminetetraacetic acid (EDTA), dipicolinic acid, or o-phenanthroline.[9]
New Delhi metallo-β-lactamases (NDM) are one of the most resistant carbapenemases first discovered in Klebsiella spp. isolated from a case of urinary tract infection in 2008 in India.[10] NDM-positive strains of E. coli and Klebsiella spp. are highly drug resistant, being sensitive only to few drugs such as tigecycline and colistin.[1112] NDM-1 has also been recovered from bacteria causing various infections such as wound and bloodstream infections from different parts of the country and worldwide.[13] It has become a major cause of concern to the clinicians as they are left with very few treatment options.
To create awareness about the carbapenemase resistance, several studies have been conducted around the world. We have planned this study to know the prevalence of blaNDM-1 gene-producing E. coli among hospitalized patients and the antimicrobial resistance pattern among these isolates in our tertiary care setup.
Materials and Methods
The present study was conducted in the Department of Microbiology, Hind Institute of Medical Sciences, Barabanki, Uttar Pradesh, India. Five-hundred nonduplicate E. coli obtained from clinical samples including pus, urine, blood, sputum, various body fluids, and wound swab received from wards of inpatient departments of the hospital during January 2014–August 2014 were included in the study.
Sample processing
Various clinical samples received in the bacteriology laboratory of the microbiology department were processed as per the standard protocol.[14] Pus, sputum, various body fluids, and wound swab were inoculated on 5% sheep blood agar and MacConkey agar and urine samples on cysteine lactose electrolyte-deficient agar media. After inoculation, all the media were incubated in an incubator aerobically at 37°C for 16–24 h.
Identification of Escherichia coli
The culture plates were examined for the growth after incubation and were processed for further identification. The identification of E. coli was done on the basis of colony morphology, Gram's staining, and biochemical tests.
Phenotypic detection of carbapenemase production
Isolates were first screened for the production of carbapenemase and then confirmed phenotypically as per CLSI 2014 guidelines.[15]
Screening test
The screening was done by disc diffusion method using ertapenem 10 μg disc. A lawn culture of 0.5 McFarland standard suspension prepared from the isolate to be tested was done on Mueller–Hinton agar (MHA) and ertapenem disc was applied on the surface of agar and incubated aerobically at 37°C for 16–18 h. The zone of inhibition between 19 and 21 mm was considered indicative of carbapenemase production. Quality control strain used for the test was E. coli ATCC 25922.
Confirmatory test
Isolates which showed positive screening test and found resistance to one or more agents in cephalosporin subclass III such as cefoperazone, cefotaxime, ceftazidime, ceftizoxime, and ceftriaxone (each with potency 30 μg/disc, HiMedia) were further subjected to confirmation by modified Hodge test (MHT). A lawn culture of 1:10 times diluted 0.5 McFarland suspension of E. coli ATCC 25922 in saline was done on MHA plate. After 3–10 min of drying, ertapenem 10 μg disc (HiMedia) was applied. Using an inoculating loop, 3–5 colonies of the test isolate along with the negative and positive control were streaked in a straight line of at least 25 mm out from the edge of the disc. The plates were then incubated at 37°C for 16–20 h. After incubation, the plates were examined for the enhancement of growth of E. coli strain at the intersection of the test or quality control isolates and the zone of inhibition. An enhanced growth is indicative of carbapenemase production. Klebsiella pneumoniae ATCC BAA-1705 was taken as MHT-positive control and K. pneumoniae ATCC BAA-1706 was taken as MHT-negative control.
Antimicrobial susceptibility profile of carbapenemase producer
Antimicrobial susceptibility profile of all the isolates showing carbapenemase production by phenotypic method was determined by Kirby–Bauer disc diffusion method as per CLSI 2014 guidelines.[15] The antibiotics used (potency in μg/disc) were ampicillin (10), gentamicin (10), tobramycin (10), amikacin (10), ceftazidime (30), ceftriaxone (30), cefotaxime (30), cefuroxime (30), cefazolin (30), cefepime (30), aztreonam (30), amoxicillin–clavulanate (10/10), piperacillin–tazobactam (100/10), ticarcillin–clavulanate (75/10), ciprofloxacin (5), levofloxacin (5), norfloxacin (10), doripenem (10), imipenem (10), meropenem (10), and nitrofurantoin (300).
Genotypic detection of New Delhi metallo-β-lactamase-1 production
DNA was extracted from the isolates by Real Genomics DNA Extraction Kit (version 2013-1, RBC Bioscience Ltd.). The extracted DNA was preserved at 4°C until polymerase chain reaction (PCR) was performed. PCR was performed to determine the blaNDM-1 gene in the carbapenemase-producing Gram-negative isolates. Primers used for the amplification of the gene were NDM-Fm (5′-GGTTTGGCGATCTGGTTTTC-3′, positions 133–153): forward primer (FP) and NDM-Rm (5′-CGGAATGGCTCATCACGATC-3′, positions 734–754): reverse primer (RP) which amplified an internal fragment of 621 bp of the blaNDM-1 gene.[16] The amplification of DNA was performed in Applied Biosystems thermocycler in a final volume of 25 μl containing 12.5 μl of Master mix (2X), 1 μl of FP (10 mM), 1 μl of RP (10 mM), 8.5 μl of Milli-Q water, and 2 μl of DNA template. The PCR conditions for the amplification of gene was comprised of denaturation at 94°C for 10 min; 36 cycles of amplification at 94°C for 30 s, 54°C for 40 s, and 72°C for 50 s; and final extension at 72°C for 5 min. The amplified product was obtained and kept at 4°C until loaded in gel for electrophoresis. The amplified DNA products were visualized by gel electrophoresis using 1% (w/v) agarose gel in 1X TBE (1.0 M Tris/0.9 M boric acid/0.01 M EDTA) with 5 μl of ethidium bromide [Figure 1].
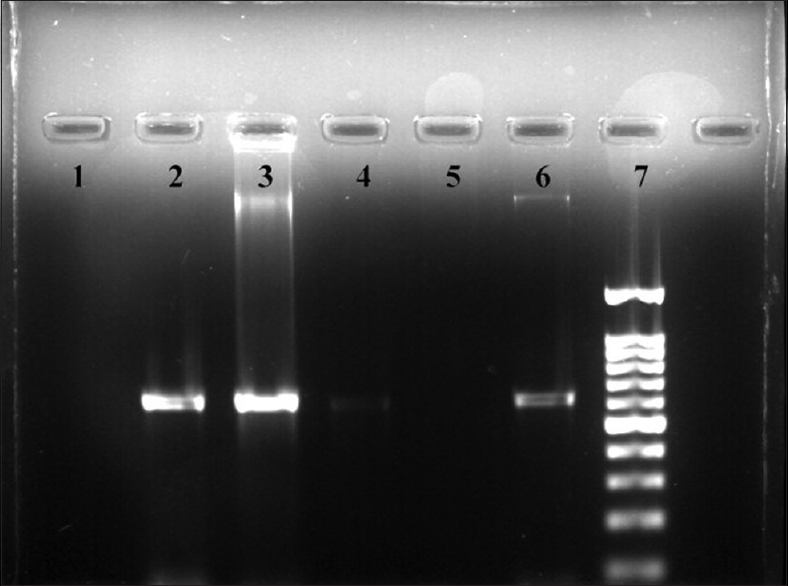
- Gel electrophoresis picture of New Delhi metallo-beta-lactamase-1 gene showing four samples
Ethics
Ethical committee approval was taken before the study and patient's detailed clinical history was recorded in a preformed questionnaire.
Statistical analysis
The present study was a prospective cross-sectional observational study conducted in the Department of Microbiology, Hind Institute of Medical Sciences, Barabanki, Uttar Pradesh, India. The results were recorded in a Microsoft Excel sheet and were analyzed statistically. Chi-square test was used to analysis the data and P < 0.05 was considered as statistically significant.
Results
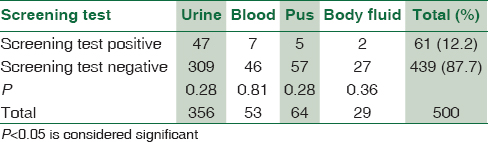
During the study, 500 nonduplicate E. coli isolates were obtained from the hospitalized patients; of these 61 (12.2%) were carbapenemase producers detected by initial screening test. Urinary tract infection was the most common infection encountered which constitutes 47 urine isolates. Other E. coli isolates were from blood (7), pus (5), and other body fluids (2) [Table 1]. From 61 E. coli isolates, 36 (7.2%) were detected positive for NDM-1 gene by conventional PCR. On all 61 E. coli isolates, MHT was performed, where 47 (9.4%) isolates confirmed carbapenemase production by phenotypic confirmatory test [Table 2]. All isolates with NDM-1 gene were MHT positive (100%). Eleven MHT-positive E. coli isolates were negative for NDM-1 gene.
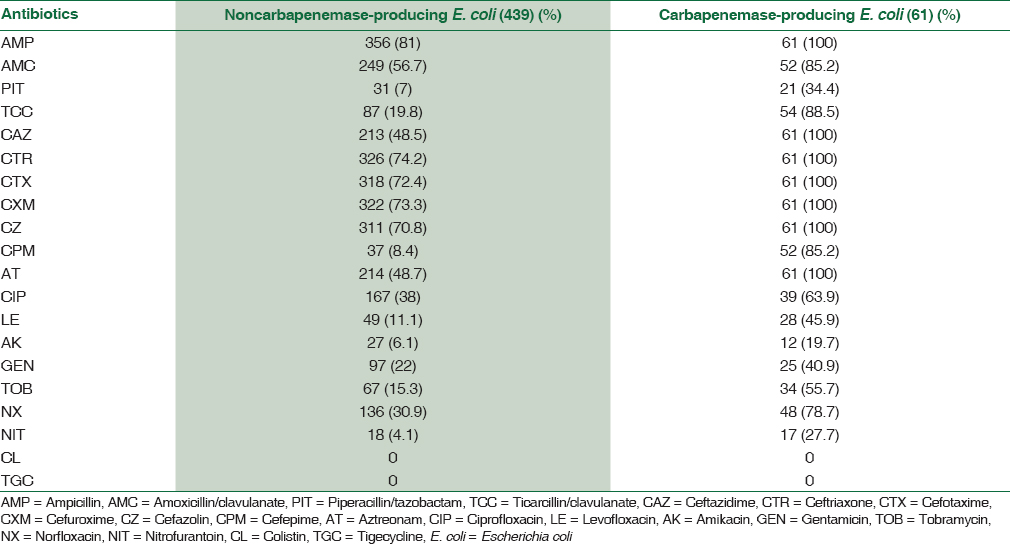
By the disc diffusion method of antibiotic sensitivity, all 61 carbapenemase-producing E. coli isolates were completely resistant to meropenem, imipenem, ertapenem, and doripenem. These isolates showed 100% resistant to ampicillin, third-generation cephalosporins, and aztreonam but variably resistant to ampicillin/clavulanate (85.2%), ticarcillin/clavulanate (88.5%), cefepime (85.2%), ciprofloxacin (63.9%), tobramycin (55.7%), levofloxacin (45.9%), gentamicin (40.9%), piperacillin/tazobactam (34.4%), and amikacin (19.7%), whereas norfloxacin (78.7%) and nitrofurantoin (27.7%) in urine with their descending order of resistance [Table 3].
Discussion
Carbapenemase resistance especially in gram negative bacteria is an emerging threat to the world of antimicrobial agents and has become a major concern to the clinicians. Increasing carbapenemase resistance among E. coli has become a challenge in the present era of antibiotic resistance. In our study on E. coli, we have taken 500 nonduplicate samples, which showed 61 carbapenemase-resistant isolates by initial screening test. The study by Khajuria et al., on urinary pathogens showed 45 carbapenemase-resistant isolates out of 300 urinary E. coli isolates by disc diffusion method and E-test.[12] In 2013, a study conducted in a surgical Intensive Care Unit (ICU) of a hospital in Egypt documented 21% (50/229) carbapenemase resistance among Gram-negative bacilli from which only one isolate was E. coli.[17] We have seen low carbapenem resistance level (n = 61; 12.2%) in our study as the isolates were taken from wards and these isolates are more prevalent in ICUs.[818] A study in Kuwait hospital in 2014 reported 8% (61/764) carbapenemase resistance among Enterobacteriaceae including 22 resistant E. coli isolates.[19]
We have seen that urinary tract infection (n = 47; 77%) was the most common infection among the carbapenemase-resistant E. coli isolates followed by bloodstream infection (n = 7; 11.5%) and pus/wound infection (n = 5; 8.2%) similar to Jamaal et al's. study in Enterobacteriaceae members.[19] In Shenoy et al's. study, respiratory tract infection and urinary tract infection were the main infections associated with carbapenemase-producing Gram-negative isolates.[20]
We have performed MHT on all the 61 screened E. coli isolates and we noted that 47 isolates were confirmed carbapenemase producers by phenotypic confirmatory test. In a study from Pakistan, 138 (69%) Gram-negative isolates were positive for MHT out of a total 200 isolates which were showing intermediate or susceptible zone, i.e., 16–21 mm for imipenem.[21]
In the present study, the prevalence of NDM-1 among all E. coli isolates is 7.2% (36/500). A report from Times of India, October 2011, stated that 8.1% NDM-1 E. coli were isolated from wards and ICUs of Sir Ganga Ram Hospital.[22] A study from Northeast detected 5.18% (14/270) prevalence of NDM-1 E. coli isolates from the tertiary care setup.[11] From the other parts of the world, the prevalence of NDM-1 gene among Enterobacteriaceae reported was 2.7% in Kuwait and 1.2% in India, Pakistan, and the United Kingdom.[1923]
NDM-1 gene accounts for 59% (36/61) of all carbapenemase-producing E. coli isolates by conventional PCR which is similar to 59 NDM-1 gene positive from 100 carbapenem resistance Gram-negative isolates through multiplex PCR where out of 59 NDM-1 gene, 14 were E. coli isolates.[24] Shenoy et al. reported 45.9% NDM-1 gene from all MDR Gram-negative bacilli.[20]
Bora A et al. and Khajuria et al. showed 100% MHT positivity with NDM-1 gene E. coli.[1112] In our study, all NDM-1 gene E. coli isolates were MHT positive, but there were 11 MHT-positive isolates which were NDM-1 gene negative. There are also studies reporting NDM-1 gene-positive strains with negative or weakly positive MHT results.[2025] The percentage of resistance to antibiotics among these 11 E. coli isolates was less as compared to NDM-1 gene E. coli isolates.
In our study, carbapenemase-producing isolates were more resistant to other antibiotics such as aminoglycosides and fluoroquinolones along with β-lactam group compared to noncarbapenemase-producing E. coli.[11] Fomda et al. from Kashmir and other studies reported nearly 100% resistance to all antibiotics (β-lactam group, aminoglycosides, tetracyclines, co-trimoxazole, and fluoroquinolones) except tigecycline and polymyxin B in NDM-1 isolates.[1726] These multidrug-resistant isolates were completely sensitive to colistin and tigecycline.[1223] There are also studies reporting the resistance to colistin and tigecycline similar to the study by Mataseje et al. showing 7% and 18.6% resistance to colistin and tigecycline, respectively.[27]
Our study was limited to NDM-1 detection, so we cannot comment about other metallo-β-lactamase and carbapenemase enzymes. Minimum Inhibitory Concentration (MIC) of carbapenems were not determined which could have given more information and clinical correlation. Follow-up of the cases with NDM-1 gene carrying E. coli isolates was done and found two deaths (5.5%), whereas rest of the patients survived.
Conclusion
The higher antibiotic resistance among carbapenemase-producing isolates is a matter of great concern for the therapeutic point of view. The carbapenemase-producing isolates still have sensitivity to colistin and tigecycline, but tigecycline is not used in urine, so in urinary pathogens, colistin remains the mainstay of treatment. Studies with resistance to colistin and tigecycline are also documented. Hence, if the present scenario continues, we may be left with no antimicrobial agents of choice for treating such deadly pathogens. Early detection can help prevent the spread of these microorganisms and a proper treatment in time reduces the risk of mortality.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- The distinct category of healthcare associated bloodstream infections. BMC Infect Dis. 2012;12:85.
- [Google Scholar]
- Escherichia, Klebsiella, Enterobacter, Serratia, Citrobacter, and Proteus. In: Medical Microbiology (4th ed). Galveston, Texas: University of Texas Medical Branch; 1996.
- [Google Scholar]
- Antimicrobial resistance and molecular epidemiology of Escherichia coli causing bloodstream infections in three hospitals in Shanghai, China. PLoS One. 2016;11:e0147740.
- [Google Scholar]
- Resistance in Gram-negative bacteria: Enterobacteriaceae. Am J Med. 2006;119:S20-8.
- [Google Scholar]
- Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25-64.
- [Google Scholar]
- Gram-negative bacteria that produce carbapenemases causing death attributed to recent foreign hospitalization. Antimicrob Agents Chemother. 2013;57:3085-91.
- [Google Scholar]
- Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50:3877-80.
- [Google Scholar]
- New Delhi Metallo-beta lactamase (NDM-1) in Enterobacteriaceae: Treatment options with carbapenems compromised. J Assoc Physicians India. 2010;58:147-9.
- [Google Scholar]
- Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969-76.
- [Google Scholar]
- Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046-54.
- [Google Scholar]
- Incidence of blaNDM-1 gene in Escherichia coli isolates at a tertiary care referral hospital in Northeast India. Indian J Med Microbiol. 2013;31:250-6.
- [Google Scholar]
- Emergence of Escherichia coli, co-producing NDM-1 and OXA-48 carbapenemases, in urinary isolates, at a tertiary care centre at central India. J Clin Diagn Res. 2014;8:DC01-4.
- [Google Scholar]
- Bailey & Scott Diagnostic Microbiology (12th ed). USA: Elsevier Science Health Science Div; 2007.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute; 2014.
- [Google Scholar]
- Emergence of Gram-negative bacilli with concomitant blaNDM-1 and blaOXA-48-like genes in Egypt. Am J Intern Med. 2017;5:1-6.
- [Google Scholar]
- Dissemination of the New Delhi metallo-β-lactamase-1 (NDM-1) among Enterobacteriaceae in a tertiary referral hospital in North India. J Antimicrob Chemother. 2011;66:1646-7.
- [Google Scholar]
- High prevalence of New Delhi metallo-β-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS One. 2016;11:e0152638.
- [Google Scholar]
- Phenotypic identification & molecular detection of bla(ndm-1) gene in multidrug resistant Gram-negative bacilli in a tertiary care centre. Indian J Med Res. 2014;139:625-31.
- [Google Scholar]
- Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3:189-93.
- [Google Scholar]
- Ganga Ram study finds high levels of superbug NDM-1. The Times Of India. 2011 Oct 05: Delhi
- [Google Scholar]
- Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597-602.
- [Google Scholar]
- Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among Gram negative bacilli. J Clin Diagn Res. 2014;8:DC23-6.
- [Google Scholar]
- Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: Report from the SENTRY antimicrobial surveillance program, 2006-2007. Antimicrob Agents Chemother. 2011;55:1274-8.
- [Google Scholar]
- NDM-1 (New Delhi metallo beta lactamase-1) producing Gram-negative bacilli: Emergence & clinical implications. Indian J Med Res. 2014;140:672-8.
- [Google Scholar]
- Results from the Canadian nosocomial infection surveillance program on carbapenemase-producing Enterobacteriaceae, 2010 to 2014. Antimicrob Agents Chemother. 2016;60:6787-94.
- [Google Scholar]





