Translate this page into:
Molecular Characterization Identifies Upstream Presence of ISAba1 to OXA Carbapenemase Genes in Carbapenem-Resistant Acinetobacter baumannii
Address for correspondence: Neetu Gupta, MBBS, MD, Department of Microbiology, Dr. D.Y. Patil Medical College, Hospital and Research Center, Dr. D.Y. Patil Vidyapeeth, Pune, Pimpri, Maharashtra 411018, India (e-mail: neetu.gupta@dpu.edu.in).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Evaluating the expression pattern of oxacillinases (OXA) carbapenemases is essential to understand the prevalence and spread of carbapenem resistance Acinetobacter baumannii.
Objectives
The aim of the study is to evaluate the presence of OXA carbapenemase genes and ISAba1 upstream to these genes in carbapenem-resistant A. baumannii clinical isolates.
Materials and Methods
A. baumannii isolated from clinical samples were phenotypically identified and antibiotics sensitivity was performed. Multiplex polymerase chain reaction (PCR) was used to detect OXA51-like gene, OXA carbapenemases genes (OXA-23-like, OXA-24-like, and OXA-58-like), and ISAba1 in carbapenem-resistant isolates.
Results
Out of 55 Acinetobacter isolates, 54 were confirmed as A. baumannii by PCR. BlaOXA-23-like gene was observed in 51 isolates of A. baumannii and none of the isolates showed the presence of blaOXA-24-like and blaOXA-58-like genes. Presence of ISAba1 upstream to OXA-23-like gene, OXA-51-like gene, and both OXA-51-like/OXA-23-like genes was observed in 51, 7, and 4 A. baumannii isolates, respectively.
Conclusion
The genetic pattern of carbapenem-resistant A. baumannii isolated in this study was unique, which should be factored for clinical protocols to manage infections caused by emerging resistant strains of A. baumannii.
Keywords
CRAB
ISAba1
OXA
Introduction
Multidrug-resistant (MDR) bacterial infections are responsible for increased mortality and morbidity in intensive care units (ICUs). Acinetobacter baumannii is one of the MDR pathogens which poses a significant risk to public health due to its high levels of resistance to currently used antibiotics.[1] Despite improvements in hospital infection control and antimicrobial stewardship, infections from A. baumannii continue to rise.[2,3] The ubiquitous nature of Acinetobacter poses a significant challenge to eradicate it from the hospital environments.[3] Due to the prevalence of variable antimicrobial susceptibility patterns in different hospitals, facility-specific surveillance studies are valuable in deciding tailored therapy to effectively control infections associated with A. baumannii. Further it is necessary to continuously isolate and monitor resistance patterns for adopting optimal clinical antimicrobial therapy protocols.
A. baumannii has acquired many genetic characters by horizontal gene transfer, which has led to increase in its virulence and resistance and making it a nosocomial pathogen of serious concern. Acinetobacter spp. are opportunistic pathogens that affect critical care units, where they are associated with pneumonia consequence to endotracheal tube intubations or tracheostomy procedures. They are also associated with bacteraemia's urinary tract infection and wound infections.[3] Multiple mechanisms of resistance such as β lactamases, modifications in porin proteins, and efflux pumps are reported in Acinetobacter spp. Enzymatic degradation by β lactamases is reported to be the most prevalent mechanism of resistance observed. Resistance to aminoglycosides and quinolones is reported to be due to mutations in gyrA and parC genes.[3] Carbapenems are the drugs of choice in the treatment of antibiotic-resistant Gram-negative organisms, however, increasing incidence of resistance to carbapenems is also reported.[4] Infections from carbapenem-resistant Acinetobacter baumannii (CRAB) are increasing globally and difficult to treat.[5]
A. baumannii contains intrinsic oxacillinases-51 (OXA-51) serine type oxacillinase which imparts natural resistance to β-lactams. Carbapenem resistance is reported to be due to serine oxacillinases (Ambler class D) and metallo β lactamases (MBLs) (Ambler class B). In the class D OXA-type beta lactamases, the five main groups (OXA-23-Like, OXA 24/40 like, OXA-58, OXA-143 like, and OXA-235 like) are identified. Resistance to carbapenem is reported to increase significantly when OXA genes are juxtaposed to insertion elements (ISAba1) with promoter functions.[6] However, the prevalence of these resistance factors in A. baumannii isolates from all geographical regions is not known. Hence this study was designed to evaluate the distribution of resistance genes (OXA-51-like, OXA-58-like, OXA-23-like, and OXA 24-like) and associated insertion elements (ISAba1) among clinical isolates of CRAB in a tertiary care hospital facility.
Materials and Methods
Study Design and Ethical Considerations
This study was a prospective observational study, which was reviewed and approved by the Institutional Ethical Committee. The study was conducted at the Department of Microbiology of a tertiary care hospital.
Inclusion Criteria
Carbapenem-resistant A. baumannii isolates obtained from various samples (blood, urine, pus, respiratory tract, CSF, etc.) of patients admitted in the various wards and ICUs of hospital were collected between November 2018 and December 2019. A total of 55 clinical isolates of A. baumannii were included in the study.
Exclusion Criteria
Carbapenem sensitive A. baumannii isolates obtained from various samples of patients were excluded.
Acinetobacter Baumannii Identification
A. baumannii isolates were identified by phenotypic test scheme including nonlactose fermenting colonies on MacConkey agar and preliminary tests (Gram staining, motility, oxidase, and growth at 44°C). All isolates after phenotypic identification were genotypically confirmed by detection of intrinsic blaOXA-51-like gene by PCR.[2,3]
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing (ABST) was performed on all A. baumannii isolates by Kirby–Bauer disk diffusion method.[3] The drugs for disk diffusion testing were in the following concentrations: amikacin (30μg), gentamicin (10μg), cotrimoxazole (25μg), cefoxitin (30μg), cefotaxime (30μg), ceftazidime (30μg), ceftriaxone (30μg), imipenem (10μg), meropenem (10μg), and piperacillin-tazobactam (100/10μg). Interpretation of zone of inhibition was done as per the Clinical and Laboratory Standards Institute guidelines.[7] MBL production was detected phenotypically using imipenem and imipenem-ethylenediaminetetraacetic acid (EDTA) disk. Zone of inhibition ≥ 7mm in imipenem-EDTA disk in comparison to imipenem alone was considered positive for MBL production.[8] Multidrug resistance (MDR) was considered if the isolates were resistant to at least one agent from three or more antimicrobial categories.[9]
Multiplex PCR
Bacterial genomic DNA extraction was performed using DNeasy Tissue kit, (QIAGEN) from 55 Acinetobacter isolates which showed resistance to carbapenem. Multiplex PCR was performed on all isolates to detect the presence of blaOXA-51-like, blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like genes, and ISAba1. The primer sequences used for amplification, as shown in ►Supplementary Table 1 (available online only), are previously reported.[10,11] The amplification conditions used were as follows: initial denaturation at 95°C for 5minutes, 35 cycles of 95°C for 30seconds, 50°C for 30seconds, and 72°C for 30seconds, and a final elongation cycle at 72°C for 7minutes. The presence of ISAba1 and OXA target genes was further investigated for the upstream presence of ISAba1 element to each OXA gene. To check this downstream primer of each OXA gene was combined with the forward primer of ISAba1 in a separate multiplex PCR.
Results
During the study period, 47 patients infected with CRAB were identified and 55 isolates of A. baumannii were obtained from these patients.
Antibiotic Sensitivity Testing
ABST revealed that all the isolates tested were resistant to imipenem, ceftazidime, ceftriaxone, cefotaxime, and cefoxitin. A higher degree of resistance to other antibiotics (gentamicin, cotrimoxazole, piperacillin-tazobactam, meropenem, and amikacin) was also observed (►Fig. 1). Most of the isolates (49/55, 89%) were observed to be MBL producers (►Fig. 1) and all the isolates were multidrug resistant (MDR).
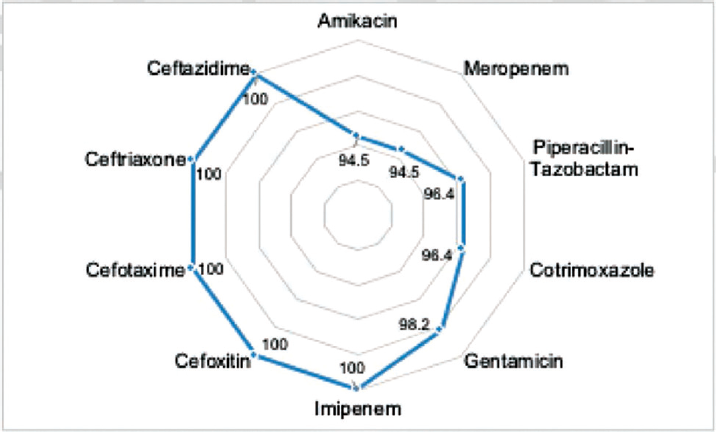
- Percentage of Acinetobacter baumannii isolates resistant to antibiotics tested.
Multiplex PCR
Out of 55 Acinetobacter isolates included in the study after the phenotypic test, 54 were confirmed as A. baumannii by PCR based on the presence of blaOXA-51 gene, which is intrinsic to this species. Of the 54 isolates, 19 were isolated from pus samples, 16 were isolated from the respiratory tract, 6 were isolated from blood samples, 4 were isolated from pleural fluid, 3 were isolated from urine samples, 1 was isolated from cerebrospinal fluid, and 5 were isolated from other clinical samples (►Fig. 2).
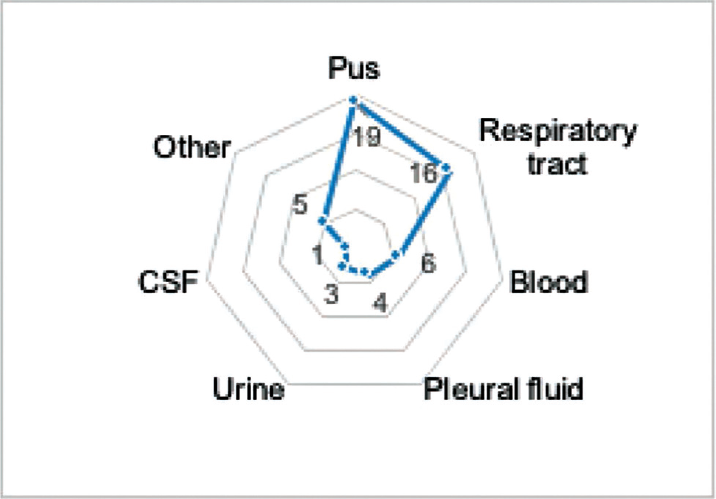
- Number of various samples used for the isolation of Acinetobacter baumannii.
Agarose gel electrophoresis (►Fig. 3) followed by multiplex PCR analysis for the presence of three carbapenemase genes (blaOXA-23-like, blaOXa-24-like, blaOXA-58-like) showed the presence of blaOXA-23-like genes in 51 isolates of A. baumannii. However, none of the isolates showed the presence of blaOXA-24-like or bla-OXA-58-like genes (►Fig. 4).
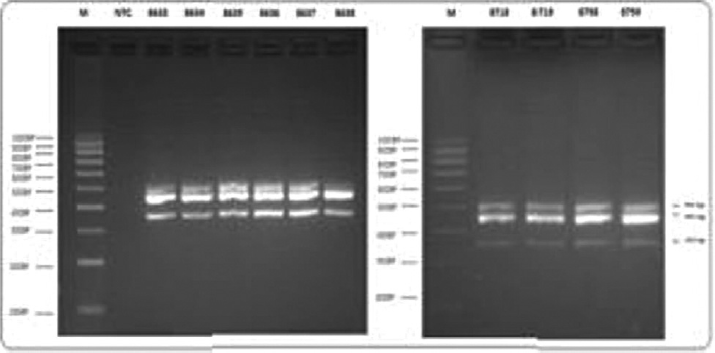
- Detection of OXA-51-like, OXA-23-like genes and ISAba1 by multiplex PCR. A representative image of agarose gel electrophoresis showing detection of genes encoding OXA-carbapenemases and ISAba1. PCR, polymerase chain reaction.

- Prevalence of OXA-carbapenemase genes in Acinetobacter baumannii isolates from various clinical samples.
Presence of ISAba1 sequence was observed in all 54 A. baumannii isolates. Fifty-one isolates showed the presence of ISAba1 upstream to OXA-23-like gene, while seven isolates detected ISAba1 upstream to OXA-51-like gene. Four A. baumannii isolates carried ISAba1 upstream to both OXA-51-like and OXA-23-like genes and interestingly three of these isolates were from pus samples and one isolate was from ET secretion (►Fig. 5).

- Prevalence and upstream presence of ISAba1 to Oxa-carbapenemase genes from various clinical samples.
In one isolate, OXA-51-like gene was not detected but the presence of OXA-23 was observed. This isolate showed ISAba1 upstream to OXA-23-like gene and was further investigated by the Vitek-2 method. The organism was identified as A. baumannii complex with excellent identification features by this method (►Fig. 5).
Discussion
Development of multidrug resistance in A. baumannii together with increasing resistance to carbapenems poses a major challenge in the treatment of patients infected with this microbe. Carbapenem resistance in A. baumannii is reported to be associated with Class D OXA carbapenemase (including intrinsic chromosomal blaOXA-51) and Class B metallo-β-lactamase production.[12] The expression of these resistance factors in addition to helping identification of A. baumannii species also contributes to increase in the minimum inhibitory concentration of carbapenem.[13] In this study a higher prevalence (94.4%) of blaOXA-23-like carrying genes was observed among the A. baumannii isolates. The OXA-23-like gene which is responsible for the development of resistance is present on the bacterial plasmid and can be transferred in conjugation between the A. baumannii strains facilitating rapid spread of resistance in any geographical regions.[14] In Asia[15] and various parts of India[16,17] the blaOXA-23-like is the dominant acquired gene detected among the CRAB. Hence our observations are consistent with these previous reports.
The blaOXA-24-like and blaOXA-58-like genes are associated with nosocomial outbreaks of MDR A. baumannii. Specifically, OXA-58 is widely reported from European Union and United States, while blaOXA-24-like is reported from United States, Spain, and China.[18,19] However, both blaOXA-24-like and blaOXA-58-like genes were not observed in any of the isolates of A. baumannii in this study. Concurrent to our observations a previous study involving five centers across India also did not detect these genes.[20] In contrast a study from South India reported OXA-24 like and OXA-58-like genes in 22.9 and 4.2% A. baumannii isolates, respectively.[21] The regional selective identification of the resistance gene, although beyond the scope of this study to explain, merits detailed evaluation in future. Expression of blaOXA-like genes is reported to play a major role in the development of carbapenem resistance, which can be potentiated by insertion sequences such as ISAba1 ISAba2, ISAba3, ISAba4, and ISAba10.[21] In this study presence of ISAba1 sequence was observed in all A. baumannii isolates. The presence of ISAba1 upstream to some OXA genes is associated with overexpression of these genes.[21] Most of the isolates in this study had upstream detection of ISAba1 to OXA-23-like gene with few isolates having ISAba1 either upstream to OXA-51-like gene or both OXA-23-like and OXA-51-like gene. Previously upstream presence of ISAba1 to OXA-23-like in Acinetobacter nosocomialis was reported from Latin America and Iran.[22,23] However, to the best of our knowledge ISAba1 expression upstream to OXA-23-like without OXA-51-like in A. baumannii/A. nosocomialis isolates is rarely reported from India. The A. baumannii isolate which did not express OXA-51-like gene but had ISAba1 upstream to OXA-23-like gene was identified as A. baumannii complex by Vitek-2 method. The variable observation of the OXA-23-like and OXA-51-like genes in A. baumannii isolates paraphs suggests intraspecies horizontal transfer of oxacillinase genes.
This study observed 100% resistance to ceftazidime, ceftriaxone, cefotaxime, and cefoxitin. The resistance pattern in this study is marginally higher than that reported by a previous study,[24] which perhaps suggests the progressive nature of resistance development by A. baumannii. Also, a majority (89%) of the A. baumannii isolates were MBL producers, indicating coexistence of both MBL and OXA-like genes in the evolving resistant strains of A. baumannii. This finding is a significant increase from 14% MBL as seen in a previous study by the same authors.[25] However, the characterization of the MBL A. baumannii isolates in this study was performed based on phenotypic and not genotypic methods, which remains the limitation of this study.
Conclusion
This study reports blaOXA-23-like gene as the predominant cause of carbapenem resistance in A. baumannii. The upstream presence of ISAba1 over blaOXA-23-like and blaOXA51-like genes, besides enhancing the development of carbapenem resistance also influences the intrastrain mobility of resistant genes. The genetic pattern of carbapenem-resistant A. baumannii isolates observed in this study should be factored for clinical protocols to manage infections caused by emerging resistant strains.
Conflict of Interest
None declared.
References
- The intensive care medicine research agenda on multidrug-resistant bacteria, antibiotics, and stewardship. Intensive Care Med. 2017;43(09):1187-1197.
- [CrossRef] [PubMed] [Google Scholar]
- Outbreak of carbapenem-resistant Acinetobacter baumannii in the intensive care unit: a multi-level strategic management approach. J Hosp Infect. 2016;92(02):194-198.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of carbapenem-resistant Acinetobacter baumannii strains isolated from hospitalized patients in Palestine. Int J Microbiol. 2017;2017 8012104
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenem-nonsusceptible gram-negative pathogens in ICU and non-ICU settings in US hospitals in 2017: a multicenter study. Open Forum Infect Dis. 2018;5(10):ofy241.
- [CrossRef] [PubMed] [Google Scholar]
- An intervention to control an ICU outbreak of carbapenem-resistant Acinetobacter baumannii: long-term impact for the ICU and hospital. Crit Care. 2018;22(01):319.
- [CrossRef] [PubMed] [Google Scholar]
- Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz J Microbiol. 2016;47(04):785-792.
- [CrossRef] [PubMed] [Google Scholar]
- CLSI document M100–S26, Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: CLSI; 2017.
- [Google Scholar]
- Metallo beta-lactamase producing pseudomonas species—a major cause of concern among hospital associated urinary tract infection. J Indian Med Assoc. 2010;108(06):344-348.
- [Google Scholar]
- Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(03):268-281.
- [CrossRef] [PubMed] [Google Scholar]
- Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(04):351-353.
- [CrossRef] [PubMed] [Google Scholar]
- Acinetobacter baumannii strains isolated from patients in intensive care units in Goiânia, Brazil: molecular and drug susceptibility profiles. PLoS One. 2017;12(05):0176790.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug resistant Acinetobacter. J Glob Infect Dis. 2010;2(03):291-304.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(10):4168-4177.
- [CrossRef] [PubMed] [Google Scholar]
- Co-existence of blaOXA-23 and blaNDM-1 genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob Resist Infect Control. 2017;6:21.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates reveals the emergence of blaOXA-23 and blaNDM-1 encoding international clones in India. Infect Genet Evol. 2019;75 103986
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect Dis Ther. 2016;5(03):379-387.
- [CrossRef] [PubMed] [Google Scholar]
- Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother. 2010;54(01):24-38.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of carbapenem-hydrolyzing class D β-lactamase genes in Acinetobacter spp isolates in China. Eur J Clin Microbiol Infect Dis. 2014;33(06):989-997.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization & epidemiology of carbapenem-resistant Acinetobacter baumannii collected across India. Indian J Med Res. 2019;149(02):240-246.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of OXA-type carbapenemase genes and genetic heterogeneity in clinical isolates of Acinetobacter spp. from Mangalore, India. Microbiol Immunol. 2011;55(04):239-246.
- [CrossRef] [PubMed] [Google Scholar]
- ISAba1/blaOXA-23-like family is the predominant cause of carbapenem resistance in Acinetobacter baumannii and Acinetobacter nosocomialis in Iran. Infect Genet Evol. 2019;71:60-66.
- [CrossRef] [PubMed] [Google Scholar]
- First report of carbapenem-resistant Acinetobacter nosocomialis isolates harboring ISAba1-blaOXA-23 genes in Latin America. J Clin Microbiol. 2013;51(08):2739-2741.
- [CrossRef] [PubMed] [Google Scholar]
- An evaluation of antibiotic profile, molecular characterization and risk factors associated with carbapenem resistant non-fermentative gram-negative isolates in a tertiary care centre. Int J Curr Microbiol Appl Sci. 2017;6(05):1057-1066.
- [CrossRef] [Google Scholar]
- Isolation and identification of Acinetobacter species with special reference to antibiotic resistance. J Nat Sci Biol Med. 2015;6(01):159-162.
- [CrossRef] [PubMed] [Google Scholar]





