Translate this page into:
Prevalence of blaKPC and its occurrence with other beta-lactamases in Klebsiella pneumoniae
Address for correspondence: Mrs. Poothakuzhiyil Remya, Department of Microbiology, Sri Ramachandra Medical College and Research Institute, Chennai - 600 116, Tamil Nadu, India. E-mail: remmoos@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Klebsiella pneumoniae (K. pneumoniae) is an important nosocomial pathogen, and the emergence of multidrug resistance in these organisms limits the treatment options for serious infections caused by them. K. pneumoniae carbapenemase (KPC) is one of the clinically significant Class A beta-lactamases.
AIM AND OBJECTIVE:
This study was aimed to detect the KPC and its coexistence with other beta-lactamases in K. pneumoniae.
MATERIALS AND METHODS:
A total of 370 isolates, collected over a period of 1 year, were included in this study. The source of these isolates were urine (n = 170), exudative specimens (n = 132), respiratory secretions such as bronchial wash, endotracheal aspirate, and pleural fluid (n = 38), and blood (n = 30). For all the isolates, antibiotic susceptibility tests by disc diffusion, modified Hodge test, and KPC screening test were done. Polymerase chain reaction (PCR) was performed for the detection of KPC and the copresence of other beta-lactamases genes.
RESULTS:
Among the 370 isolates, 41 were resistant to the carbapenem by disc diffusion and minimum inhibitory concentration tests. Screen test using ertapenem and the boronic acid disk was positive in 14 isolates. Only one isolate harbored KPC gene by PCR, and it was co-produced with SHV-12 and CTX-M-15.
CONCLUSION:
PCR remains the gold standard for detection of KPC compared with any other phenotypic methods. Early detection of these genes helps in initiating proper antibiotic treatment.
Keywords
Extended-spectrum beta-lactamases
Klebsiella pneumoniae carbapenemase
Klebsiella pneumoniae
molecular methods
phenotypic methods
Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the most clinically important Gram-negative bacilli, with high degree of multidrug resistance.[12] Carbapenems are the last resort of antibiotics used to treat serious infections caused by them. Carbapenems are dipolar and rapidly enter Gram-negative bacterial cell wall through outer membrane proteins (or porins) and target bacterial penicillin-binding proteins.[3] Carbapenemases are enzymes that inhibit almost all beta-lactam antibiotics including carbapenems and belong to the Ambler Class A, B, and D beta-lactamases.[45]
K. pneumoniae carbapenemase (KPC) is plasmid encoded and belongs to Ambler Class A enzymes.[6] Their occurrence in K. pneumoniae was first reported in 1996 from the USA. Within few years, KPC producers spread worldwide and are now encountered in many Gram-negative bacteria.[4] Of late, the coexistence of KPC with other carbapenemases in K. pneumoniae is also being encountered.[7]
From India, KPC in K. pneumoniae was first reported in 2011.[8] Since then, there has been scarcely any publication on the prevalence of KPC.[91011] Hence, this study was undertaken to detect the presence of KPC in clinical isolates of K. pneumoniae in a tertiary care hospital.
Materials and Methods
Bacterial isolates
The study included 370 clinically significant, consecutive, nonduplicate isolates of K. pneumoniae, collected over a period of 1 year (August 2014 to July 2015). The source of isolates were urine (n = 170), exudative specimens (n = 132), respiratory secretions such as bronchial wash, endotracheal aspirate, and pleural fluid (n = 38), and blood (n = 30).
Antibiotic susceptibility testing
Antibiotic susceptibility testing was done by Kirby–Bauer disc diffusion method as per the Clinical and Laboratory Standards Institute guidelines.[12] The antibiotics tested were cefotaxime (30 μg), ceftazidime (30 μg), amikacin (30 μg), ciprofloxacin (5 μg), piperacillin/tazobactam (100 μg/10 μg), and imipenem (10 μg) (HiMedia Laboratories, Mumbai). ATCC Escherichia coli 25922 was used as control strain. Determination of Minimum inhibitory concentration (MIC) to imipenem (Sigma-Aldrich, India) was detected by agar dilution method (range-0.03-128 μg/ml) according to CLSI guidelines.[12]
Phenotypic screening test
Modified Hodge test
Carbapenemase screening for the isolates was performed by the modified Hodge test (MHT).[12] ATCC Escherichia coli 25922 was cultured overnight and suspended to achieve 0.5 McFarland standard turbidity and was lawn cultured on to a Mueller-Hinton agar (HiMedia, India) plate using a sterile cotton swab. After drying, a disk containing imipenem (10 μg) was then placed at the center of the plate, and an overnight cultured test strain was heavily streaked from the center to the periphery of the plate. The presence of a distorted zone after overnight incubation was interpreted as a positive result.
Combined disk test with boronic acid
Screening for KPC-possessing isolates was done using boronic acid disk test.[13] About 120 μg of phenylboronic acid (benzeneboronic acid; Sigma-Aldrich, India) was dissolved in 3 ml of dimethyl sulfoxide and 3 ml of sterile distilled water. From this solution, 20 μl (containing 400 μg boronic acid) was dispensed onto 10 μg of ertapenem disks. Disks were allowed to dry for 30 min and used immediately or stored in airtight vials at −80°C. The test isolate was inoculated on a Mueller-Hinton agar plate as for routine Kirby–Bauer disk diffusion test, and then, disks containing ertapenem with and without boronic acid were placed on the agar. The agar plates were incubated at 37°C overnight. A ≥5-mm increase in the inhibition zone around an ertapenem disk with boronic acid compared to ertapenem disk alone was considered as positive for KPC production.
Detection of beta-lactamase-encoding genes
blaKPC gene by polymerase chain reaction
Template DNA was extracted from the isolates by boiling method.[14] All the isolates were tested for blaKPC gene using the previously described primers.[15] The primers were KPC forward-CGTCTAGTTCTGCTGTCTTG and KPC reverse-CTTGTCATCCTTGTTAGGCG. Amplicon size of KPC gene was 798 bp. Polymerase chain reaction (PCR) was performed with a final volume of 25 μl. Each reaction contained 10 pmol of each primer (Sigma-Aldrich, India), 10 mM of dNTP mixture (Takara, India), and 5U Taq polymerase (Takara, India) in 2.5 μl of 10X Taq polymerase buffer (Mg2+plus). About 2 μl of template DNA was added to 23 μl of the master mixture. Negative controls were PCR mixture with water (instead of template DNA) was included in every PCR run.
Amplification reactions were performed under the following conditions: initial denaturation 10 min at 94°C, followed by 36 cycles of amplification for 30 s at 94°C, annealing at 52°C for 40 s, and 50 s at 72°C with final extension for 5 min at 72°C. DNA fragments were analyzed by electrophoresis in a 2% agarose gel containing ethidium bromide at 100 V in IX TBE buffer.
Other beta-lactamase genes
Some of the other commonly occurring beta-lactamases were also looked for employing PCR. They were plasmid-mediated AmpC beta-lactamases and metallo-beta-lactamases such as IMP, VIM, and NDM. Class D carbapenemase such as OXA-48 and extended-spectrum beta-lactamases (ESBLs) such as SHV, TEM, and CTX-M were also tested using primers described previously.[141516] The primers used are given in Table 1.

DNA Sequencing
PCR-positive amplicons were purified and sequenced by BigDye 3.1 cycle sequencing kit using the Sanger AB13730 XL DNA analyzing instrument (SciGenom, Kerala, India). The nucleotide sequences analyzed are compared with the sequence available at the National Center for Biotechnology Information website (www.ncbi.nIm.nih.gov). GenBank accession number is KY364013.
Results
Of the 370 study isolates, 41 (11.1%) were resistant to imipenem by both disc diffusion method and minimum inhibitory concentration (MIC) determination. MIC50 and MIC90 were 0.125 μg/mL and 4 μg/mL, respectively.
Susceptibility to other classes of antimicrobials were amikacin (82.16%), piperacillin/tazobactam (80.27%), ciprofloxacin (62.97%), ceftazidime (50.54%), and cefotaxime (41.62%).
Screening for carbapenemase production by MHT was positive in 90 out of 370 isolates. Of the total isolates, KPC screening test using ertapenem and boronic acid was showed positive in 14 isolates.
blaKPC was detected in one isolate only [Figure 1]. The MIC of this isolate to imipenem was 64 μg/mL. MHT and KPC screen test were also positive. The source of the isolate was urine. This isolate also harbored blaSHV and blaCTX-M. Notably, the other beta-lactamases looked for, namely, TEM, AmpC, and MBL including NDM, IMP, and VIM, OXA-48 were not detected in this KPC-harboring isolate.
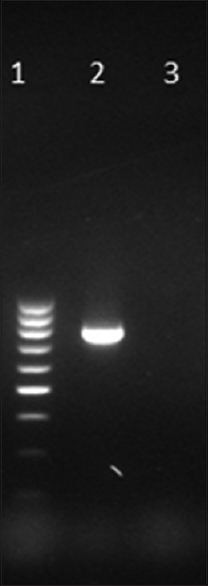
- Gel picture of amplified KPC gene. Lane 1: 100 bp DNA ladder, Lane 2: Test positive for blaKPC at 798 bp, Lane 3: Negative control
Discussion
The Ambler Class A carbapenemase was named after the organism from which it was first identified.[4] KPC beta-lactamase can hydrolyze all beta-lactams, including carbapenems, cephalosporins, cephamycins, monobactams, and clavulanic acid combinations.[17] In addition, they exhibit resistance to aminoglycosides and fluoroquinolones.[18] KPCs may not confer frank resistance to carbapenems but may impart reduced susceptibility to carbapenems. For a full blank resistance to carbapenems, impaired outer membrane permeability is often required.[19] KPC has been detected in other Gram-negative bacteria such as the family Enterobacteriaceae (E. coli, Klebsiella oxytoca, Enterobacter spp., Serratia spp., and Salmonella spp., and Citrobacter freundii) as well as in glucose nonfermenters (Acinetobacter spp. and Pseudomonas aeruginosa).[620]
Following its first detection in 1996, KPC has been reported globally. The endemic spread of KPC-producing K. pneumoniae has been reported from the USA, China, Italy, Poland, Greece, Israel, Brazil, Argentina, Colombia, and Taiwan. Sporadic occurrence has also been observed in many European countries and in several countries in the Asia-Pacific region, including India, South Korea, and Australia.[7]
The coexistence of KPC with other carbapenemase such as NDM, IMP, and VIM is common.[7] In India, the first report of KPC along with NDM-1, CTX-M-15, and SHV-12 was published in 2011.[8] Since then, there are only a few reports of KPC occurring along with other beta-lactamases.[1021] In Chennai, India, it has been observed that of the 46 carbapenem-resistant Enterobacteriaceae, 31 isolates carry blaKPC.[9] Elsewhere among 100 carbapenem-resistant Enterobacteriaceae, KPC along with NDM was detected in 14%, while KPC alone was detected in 1%.[10] Co-carriage of blaKPC along with blaNDM-1 has been reported in P. aeruginosa also.[21]
In a study conducted in Manipal, India, KPC was observed in 3.7% of 54 carbapenem-resistant K. pneumoniae.[11]
Most of the KPC-associated infections are either systemic infections, occurring in patients with multiple invasive devices or urinary tract infections without an indwelling catheter, particularly in immunocompromised patients.[19]
In this study, even though there were 14 isolates which exhibited a positive KPC screen test using boronic acid, only one isolate among them harbored blaKPC gene. The false-positive results may be due to the production of AmpC beta-lactamases or some CTX-M beta-lactamases because boronic acid derivatives are potent inhibitors of these enzymes also.[22] Thus far, there is hardly any report of KPC-2 along with SHV-12 and CTX-M-15 in a single isolate.[8]
KPC along with ESBLs limits the therapeutic options to patients, and the death rates attributed to KPC infections are high (≥50%).[23] Antibiotic options are generally limited to tigecycline and polymyxins for KPC-producing isolates.
Conclusion
Detection of KPCs using PCR remains the gold standard, since all other phenotypic tests are prone to give false-positive results. A single strain harboring multiple resistant genes is really challenging in the clinical laboratory in terms of detection and management.
Financial support and sponsorship
This study was financially supported by Founder Chancellor Ramasamy Udayar Fellowship provided by Sri Ramachandra Medical College and Research Institute.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
All the authors are grateful for the financial assistance provided by Sri Ramachandra Medical College and Research Institute as Founder Chancellor Fellowship.
References
- Sequencing and genetic variation of multidrug resistance plasmids in Klebsiella pneumoniae. PLoS One. 2010;5:e10141.
- [Google Scholar]
- Detection of Klebsiella pneumoniae carbapenemases (KPCs) among ESBL/MBL producing clinical isolates of Klebsiella pneumoniae. Int J Curr Microbiol Appl Sci. 2015;4:726-31.
- [Google Scholar]
- Status report on carbapenemases: Challenges and prospects. Expert Rev Anti Infect Ther. 2011;9:555-70.
- [Google Scholar]
- The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821-30.
- [Google Scholar]
- A structural, epidemiological & genetic overview of Klebsiella pneumoniae carbapenemases (KPCs) Indian J Med Res. 2016;144:21-31.
- [Google Scholar]
- Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895.
- [Google Scholar]
- Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother. 2012;67:243-4.
- [Google Scholar]
- BlaKPC gene detection in clinical isolates of carbapenem resistant Enterobacteriaceae in a tertiary care hospital. J Clin Diagn Res. 2013;7:2736-8.
- [Google Scholar]
- Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among gram negative bacilli. J Clin Diagn Res. 2014;8:DC23-6.
- [Google Scholar]
- Approach to carbapenemase detection in Klebsiella pneumoniae in routine diagnostic laboratories. J Clin Diagn Res. 2016;10:DC24-7.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: 25th informational supplement document M100-S25. Wayne: Clinical and Laboratory Standards Institute; 2015.
- [Google Scholar]
- Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J Clin Microbiol. 2009;47:362-7.
- [Google Scholar]
- Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153-62.
- [Google Scholar]
- Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119-23.
- [Google Scholar]
- Detection of extended spectrum beta-lactamases-producing isolates and effect of AmpC overlapping. J Infect Dev Ctries. 2013;7:618-29.
- [Google Scholar]
- Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014;22:686-96.
- [Google Scholar]
- Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin Infect Dis. 2011;53:60-7.
- [Google Scholar]
- The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228-36.
- [Google Scholar]
- Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008-2011. PLoS One. 2014;9:e116421.
- [Google Scholar]
- Co-carriage of blaKPC-2 and blaNDM-1 in clinical isolates of Pseudomonas aeruginosa associated with hospital infections from India. PLoS One. 2015;10:e0145823.
- [Google Scholar]
- Inhibitor-based methods for the detection of KPC carbapenemase-producing Enterobacteriaceae in clinical practice by using boronic acid compounds. J Antimicrob Chemother. 2010;65:1319-21.
- [Google Scholar]
- Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791-8.
- [Google Scholar]





