Translate this page into:
Virulence determinant and extended spectrum beta-lactamase production in Klebsiella pneumoniae isolated from a tertiary care hospital, South India
Address for correspondence: Mrs. Lathamani Kotekani, Research Scholar, Department of Microbiology, KVG Medical College, SULLIA, Dakshina Kannada, Karnataka, India. E-mail: lathamani2006@rediffmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Klebsiella pneumoniae on these days show greater resistance towards newer generation cephalosporin. The present study made an effort to show the relevance between ESBL producing K. pneumoniae and virulence determinant in respect to serum resistance and K1, K2 antigens at a tertiary care hospital in South India.
MATERIAL AND METHODS:
A total of 520 consecutive, non-duplicate isolates of K. pneumoniae were recovered from various clinical specimens such as Urine (n = 360), sputum (n = 107), pus (n = 25), blood (n = 6) and other miscellaneous specimens (n=22) obtained from both out patients and in patients between June 2012 to July 2016 were included in the study.
RESULTS:
Polymerase Chain Reaction to detect bla genes in 62 isolates phenotypically identified as ESBL producers were successful in 58 (93.54%) isolates. Of the 13 ESBL producing hypermucoviscous Klebsiella pneumoniae (hvKP) strains, 6 of them were showing the amplicons for gene's coding for K1 antigens.
CONCLUSION:
The study provides further authentication of the global sporadic of bla CTX-M and the relevance between K antigens and serum resistance with ESBL production in our place. As there is no much study available, it also highlights the need for further study of their epidemiological surveillance.
Keywords
BlaCTX-M
blaSHV
blaTEM
extended spectrum beta-lactamase
Klebsiella pneumoniae
Introduction
Klebsiella pneumoniae is the most often isolated Klebsiella species from human infections and is responsible for wide variety of infections, including septicemia, wound infections, urinary tract infections, cholecystitis, and pneumonia (Friedlander's disease).[1] The spread of K. pneumoniae infection is more of an epidemic than endemic nature and responsible for pandemics of diseases due to indiscriminate usage of antibiotics.[2] K. pneumoniae on these days show greater resistance toward newer generation cephalosporins.[3] Extended-spectrum beta-lactamases are plasmid mediated enzymes that capable of hydrolyzing beta-lactam antibiotics such as penicillins, oxyimino-cephalosporin, and monobactam.[4] The beta-lactam inhibitors such as clavulanic acid, tazobactam, and sulbactam have limited antibacterial activity of its own, but when they combine with beta-lactam antibiotics, they block the beta-lactamase activity and render the drugs active. The double-disk synergy test is one of the phenotypic testing of extended spectrum beta-lactamase (ESBLs) which is an incomplete method to find ESBLs due to its less sensitivity and specificity. Therefore, the present study has opted polymerase chain reaction (PCR) to detect bla genes among K. pneumoniae. The CTX-M ESBLs were rare enough to be noticed until 1990s. However, in recent years, there is a drastic change in the epidemiology of ESBLs with the worldwide emergence and spread of the CTX-M-type enzyme easily surpassing those of blaSHV and blaTEM ESBL genes.[5] Based on CTX-M amino acid similarities, there are five major groups of blaCTX-M such as 1, 2, 8, 9, and 25. Occurrence of blaCTX-M genotype varies geographically. A subtype of CTX-M 1, the blaCTX-M-15, described first in Escherichia coli and isolated from India during 2001 has now disseminated to all continents.[67] Hence, the present study planned to characterize the dominant enzyme variants such as TEM, SHV, and CTX. Studies have shown that K1 or K2 capsular serotypes and hypermucoviscous (HMV) phenotypes are responsible for the virulence determinants of K. pneumoniae infections.[8]
Many smooth strains of Gram-negative bacteria show the bactericidal effect toward normal human serum and this effect may serve as an important factor to cause some of the infections.[9] Serum resistance is the virulence property of an organism by which they escape the lytic action of complement system of the normal serum.[10] Studies have shown that strains of Klebsiella producing extended-spectrum beta-lactamases are significantly more invasive with more fimbrial adhesions and more resistant to the normal human serum bactericidal effect than nonESBL-producing strains.[11]
To understand the problem of choice of antibiotics and effective treatment with antibacterial substances in the population, there is a need of the surveillance program toward antibacterial resistance. The present study will help out to know the antibacterial resistance pattern in respect to third generation cephalosporin among the isolates of K. pneumoniae with production of ESBLs in Dakshina Kannada District with respect to 3 major types of ESBL genes and to know the relevance between ESBL-producing HMV K. pneumoniae (hvKP) and K1, K2 antigens. The study also shows the occurrence of serum resistance and its relevance with ESBL production among the isolates studied.
Materials and Methods
The proposed study was carried out in the Department of Microbiology, KVG Medical College and Hospital, Sullia, Dakshina Kannada, Karnataka, India. A total of 520 consecutive, nonduplicate isolates of K. pneumoniae were recovered from various clinical specimens such as Urine (n = 360), sputum (n = 107), pus (n = 25), blood (n = 6), and other miscellaneous specimens (n = 22) obtained from both outpatients and in patients between June 2012 and July 2016 were included in the study. The identification of the strain was carried out by standard microbiological techniques.[12]
Antimicrobial susceptibility testing
Antibacterial susceptibility of all the isolates of K. pneumoniae was determined by the standard Kirby-Bauer disc diffusion method as per the guidelines established by CLSI.[13] The antimicrobial disks used in the present study were ciprofloxacin (5 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), amikacin (30 μg), gentamicin (10 μg), imipenem (10 μg) ertapenem (10 μg), ampicillin (10 μg), and piperacillin/tazobactam (100/10 μg).
Detection of extended-spectrum beta-lactamase production
Screening of extended-spectrum beta-lactamase production
Disk diffusion test was used to know the resistance to third-generation cephalosporin such as cefotaxime, ceftazidime, and ceftriaxone as per the CLSI guidelines.[13] The antibiotic disks, such as cefotaxime, ceftazidime, and ceftriaxone were kept on the surface of the media and the plates incubated in air at 37°C for 18–24 h. A positive screening test for ESBL production was to develop resistance to at least one of the above antibiotics used for the study.
Confirmation of extended-spectrum beta-lactamase detection by double disc synergy test
The production of ESBL by the strains was determined by a screening test, double-disc synergy test described by Jarlier et al.[14] An augmentation of zones of tested third generation cephalosporin by clavulanic acid indicated the possible existence of an ESBL.
Detection of bla genes by polymerase chain reaction
For DNA extraction, 62 of the randomly selected isolates exhibiting ESBL production in the initial screening test were made use of as per the procedure given by Bora et al.[15] Procedure was slightly modified to get the largest amount of DNA.
Five to six colonies of K. pneumoniae suspended in 200 μl double-distilled water and heated at 95°C for 10 min and added with equal proportion of 90% ethanol to DNA to get precipitated. The precipitated top solution was again added with equal proportion of 90% ethanol and centrifuged at 6440 × g for 5 min. Discarded the supernatant and to the deposit again added equal proportion of 90% ethanol and repeated the procedure for two more times. At the end of the procedure, the deposit was left to dry and added double distilled water to remove ethanol and again centrifuged at 6440 × g for 5 min. The deposit with few drops of nuclease-free water was directly used as template DNA for PCR study. The primers used and PCR cycling conditions were described in [Tables 1 and 2].

The total concentration of PCR reaction to do single test was 25 μl [PCR ready mix-12.5 μl (Sigma Aldrich, Merck's Life Science), Forward primer-1 μl, Reverse primer-1 μl (Bioserve Hyderabad, India), Template DNA-4 μl and Nuclease free water-6.5 μl (Hi Media Laboratories Pvt, Ltd., Mumbai, India)].
Controls used
Positive control: For blaSHV-K. pneumonia ATCC 700603 (courtesy of Dr. Srikanth, HOD, Dept. of Microbiology, St. John's Medical College, Bangalore, India). For blaTEM and blaCTX-M genes-a previously confirmed K. pneumoniae isolate possessing blaTEM and blaCTX-M genes.
Negative control-nuclease-free water Uniplex PCR was made use to detect blaTEM, blaSHV and blaCTX-M-1 genes. The samples that were showing positive amplicons for the genes coding for CTX-M group-1 gene were again processed for the detection of PCR amplification of blaCTX-M-15 gene by uniplex PCR. A multiplex PCR was processed to detect the genes coding for CTX-M groups-2, 8, 9, and 25 genes.
Thermocycler (Quanta Biotech, 96 S) was used to run PCR cycles; PCR run conditions were programmed using the supplied software.
Post-PCR validation was performed by gel electrophoresis, and the ethidium bromide stained PCR products were visualized using Gel Doc system under UV light.
String test
HMV is a phenotypic property of K. pneumoniae. The present study made use of a string test to confirm it. A loop was used to stretch the colonies of K. pneumoniae grown on Lauria Bertani medium at 37°C. Viscous strings of >5 mm in length was a positive test.[21]
Serum resistance
The cell suspension of fresh K. pneumoniae grown in blood agar by adjusting the turbidity standard, 0.5 McFarland (1.5 × 108 cfu/ml) was made to suspend in Hank's balanced salt solution. At about 0.05 mL each of suspension of K. pneumoniae and fresh undiluted healthy human serum was made to incubate at 37°C for 15 min. Each ten microliters of the above preparation were spread over the blood agar plate. The viable bacterial colony was made to count at 0 min and after 180 min of incubation. The bacteria read as serum sensitive if there was drop in viable count to 1% of the initial value and serum resistant if >90% of K. pneumoniae isolates survived after 180 min.[22]
Results
The antimicrobial susceptibility pattern of clinical isolates of K.pneumoniae studied in respect to the third generation cephalosporin wasshown in Table 3. Out of 520 isolates of K. pneumoniae tested, 316 (60.76%) of them were showing resistance to third-generation cephalosporin. One hundred and forty-two (44.93%) was found to produce ESBLs using phenotypic detection method.

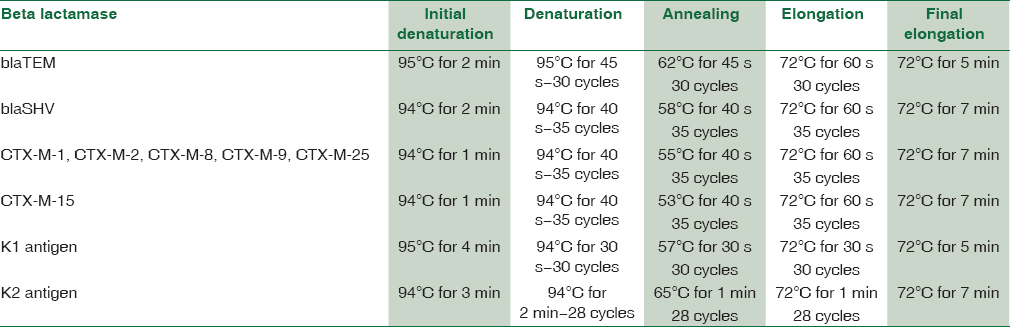
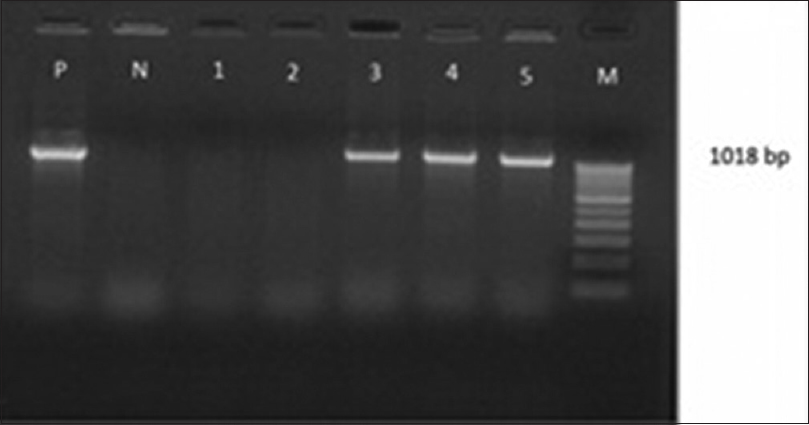
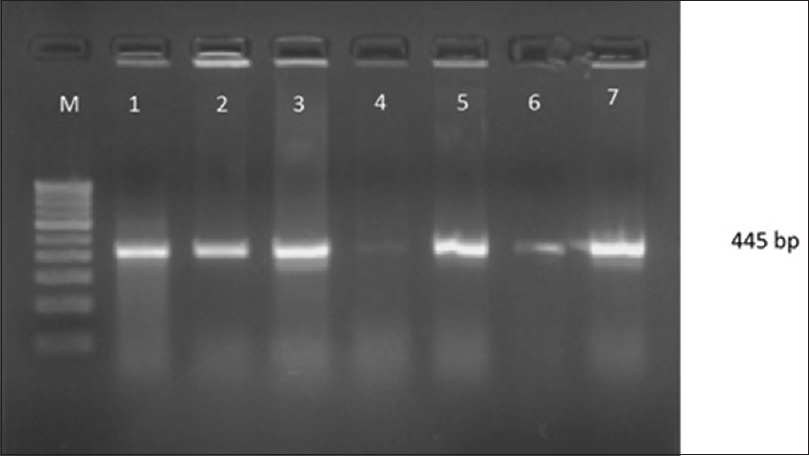
Attempts to detect one or more bla genes by PCR in 62 randomly selected isolates phenotypically identified as ESBL producers were successful in 58 (93.54%) isolates. Out of 62 isolates of K.pneumoniae studied for PCR to detect bla genes, 45(77.58%) harbored blaSHV gene [Figure 1]. 49(84.48%) blaCTX-M-1 gene [Figure 2], Only 2 (15.38%) were showing positive genes for CTX-M-9 out of 13 CTX-M-1 and SHV negative strains tested [Figure 3]. CTX-M-15, a sub type of CTX-M-1 was found in 37 (75.51%) of 49 CTX-M-1 positive strains [Figure 4]. None of them was harboring TEM, CTX-M-2, CTX-M-8 and CTX-M-25 bla genes among the tested strains of K.pneumoniae. It has been also observed that 38 (61.29%) of them were harboring both CTX-M and SHV genes.
- Gel Doc picture showing polymerase chain reaction-amplified product of bla SHV. Lane M – 1 Kb DNA ladder, Lane 3-5 bla SHV positive amplicons (1018 bp), Lane 1-2- Isolates of Klebsiella pneumoniae without bla SHV, Lane N-Negative control, Lane P-Positive control
- Gel Doc picture showing polymerase chain reaction amplified product of blaCTX-M-1. Lane M- marker (100bp DNA ladder), Lane-1-7- positive amplicons of clinical isolates of Klebsiella pneumoniae with blaCTX-M-1 (445 bp)
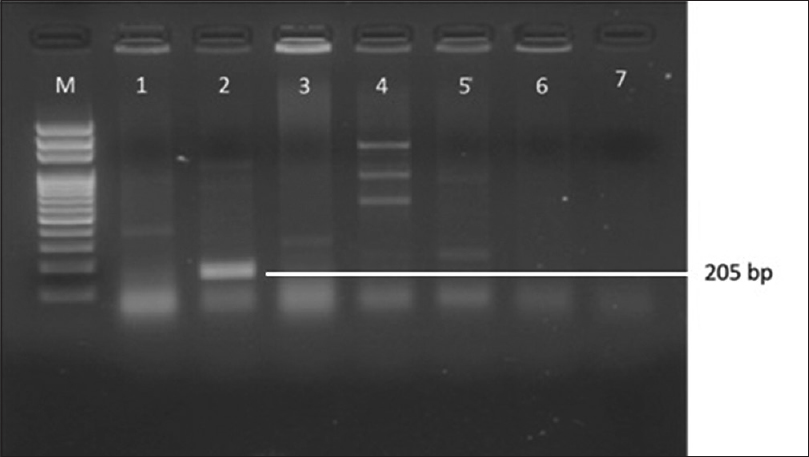
- Gel Doc picture showing CTX-M - 9 polymerase chain reaction product on a 2% Agarose gel. M- marker (100bp DNA ladder), Lane 2 and 5 - bla CTX-M-9 positive amplicons (205 bp)
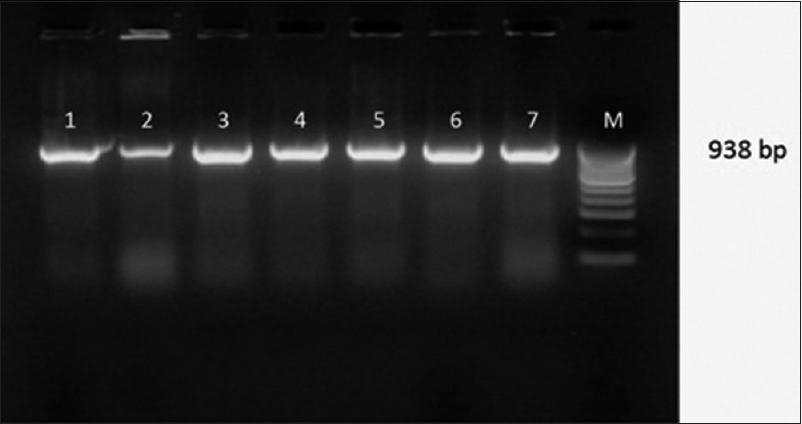
- Gel Doc picture showing CTX-M -15 Uniplex polymerase chain reaction products on a 2% Agarose gel. Lane 1,2,3,4,5,6,7- clinical isolates showing bla CTX-M-15 positive amplicons (938bp), M- marker (100bp DNA ladder)
The string test was found positive among 13 of 62 (20.96%) ESBL strains of K.pneumoniae tested. Of the 13 string test positive strains, 6 of them were showing the amplicons for genes coding for K1 antigens [Figure 5]. None of them were harboring the genes coding for K2 antigen.
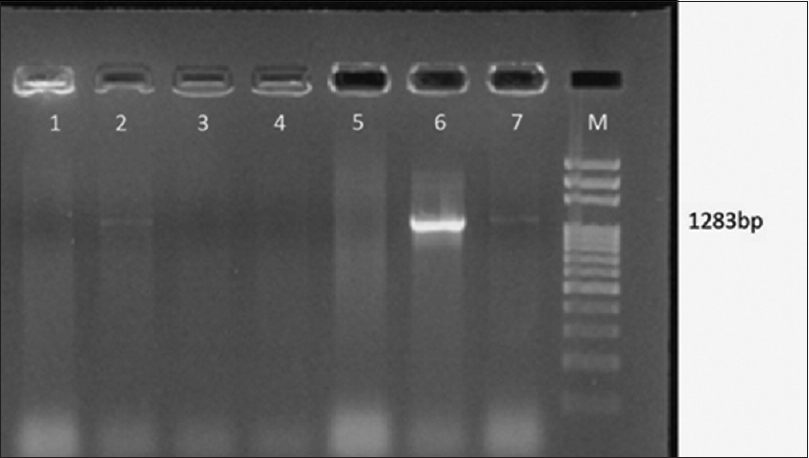
- Gel Doc picture showing polymerase chain reaction amplified product of genes for K1 antigen. Lane M – 1 Kb DNA ladder, Lane 2, 6, 7 positive amplicon genes for K1 antigen (1283 bp) of isolates of Klebsiella pneumoniae
Out of a total of 520 clinical isolates of K. pneumoniae, 232 (44.6%) showed the property of serum resistance and of these 108 (76.05%) ESBL producers were positive for serum resistance.
Discussion
Although there are many studies with a vast amount of data from different parts of the country, they differ widely in study period, patient population, sample type and size, detection method. Hence, it is found difficult to interpret the result meaningfully. The highest prevalence rate of ESBL production among K. pneumonia was primarily found in Latin America; where about 50% of them harbor ESBLs.[2324] The data collected before 2000 showed that, 96% ESBL producers among K. pneumoniae were prevalent in five cities of India.[25] Extended-spectrum beta-lactamase exhibit low-level resistance in vitro. Therefore, routine disc susceptibility tests done in the laboratories may falsely interpret as sensitive for ESBL production among bacterial isolates.[26]
In the present study, out of the 316 resistant strains of K. pneumoniae toward third-generation cephalosporin, 142 (44.93%) was found to produce ESBLs using phenotypic detection method. In contrast to the CLSI phenotypic confirmatory test for ESBL production, genotypic detection was found to positive only for 58/62 (93.54%) of K. pneumoniae isolates in this study. Multiple factors contribute to decide the phenotypic confirmatory tests for ESBL detection to be falsely positive or negative. This includes, lower sensitivity of phenotypic method, occurrence of resistance due to the influence of environmental factors, varying substrate affinities, the inoculums effect, multiple different β-lactamase types, and ESBL production by an individual isolate of a bacteria that fundamentally produce the AmpC β-lactamases.[2728] The amplification of CTX-M-specific gene products give much more evidence that a blaCTX-M gene is responsible for ESBL production in an isolate without further sequencing.[29] The PCR data using CTX-M group-specific primers of our study revealed a remarkable increase in the CTX-M-1 genotype (49 [84.48%]). Similar report was made in different parts of India.[30313233] Meanwhile, 45 (77.58%) of ESBL strains were harboring SHV genes. A study from the Northern part of India revealed 72% of bla SHV production.[34] Whereas studies from Southern part of India showed 0% SHV and bla TEM genes production.[35] In our study, there were no specific amplicons detected among the ESBL strains of K. pneumoniae tested samples for blaTEM. Hence, the result reported was 0%. CTX-M-8 and CTX-M-9 groups have been rarely reported from India.[36] Our study reported 2 (15.38%) of ESBL strains tested belonged to CTX-M-9 cluster. The spread of CTX-M-9 cluster of genes was also reported from Brazil.[37] Further, study of PCR using CTX-M-15 primer showed the highest occurrence 37 (75.51%) of CTX-M-15 genes among the CTX-M-1 positive strains. Many other researchers reported similar results.[3839] Many other studies have shown that the CTX-M gene showed its highest predominance among Klebsiella spp. and are responsible for sporadic.[4041] There was no specific amplicons found for CTX-M-2, CTX-M-8, and CTX-M-25 bla genes among the tested strains of K. pneumoniae. The CTX-M group is a new family of plasmid-mediated ESBL, and now, it is the most prevalent and predominant ESBL encoding gene found worldwide replacing TEM and SHV types in many European and Asian countries similarly.[4243]
The ESBL producers of K. pneumoniae usually carry multiple resistant genes to provide them to persist. The present study revealed that the 36 (64.28%) of the strains tested coexist bla SHV and bla CTX-M genes. Many other researchers reported the same.[4445] Our study showed that, the string test was positive among 13 of 62 (20.96%) ESBL strains of K. pneumoniae tested. Of the 13 string test-positive strains of hvKP, 6 of them were showing the amplicons for genes coding for K1 antigens. None of them were harboring the gene's coding for K2 antigens. However, the study conducted by Jenney et al. revealed that, out of 293 isolates of K. pneumoniae studied for serotyping, 6 of them belonged to K1 antigens and 4 of them of K2 antigens. However, none of the K1 and K2 positive isolates were ESBL producers.[46] In our study, total number of strains showing serum resistance was 232 (44.6%). The rate of serum resistance among ESBL producers was 108/142 (76.05%). However, in non-ESBL producers, it was 86/174 (49.42%). To agree the present study, Sahly et al. found that, Klebsiella strains producing ESBL showed higher percentage (30%) of resistance toward serum bactericidal effect than non-ESBL producing Klebsiella strains (17.9%). Knowledge of the resistance pattern of the strains, ESBL production in bacterial strains and restricted use of antibiotics in a geographical area will help us to lead proper and judicial use of antibiotics to relieve the selective pressure put by unlimited use of antibiotics.
Conclusion
In conclusion, this study has highlighted the high percentage of ESBL producers among the clinical isolates of K. pneumoniae in South Kanara District. The routine antibiotic susceptibility test done in the laboratory erroneously detects isolates sometimes sensitive to any of the third generation cephalosporins. Hence, it is mandatory to detect ESBL production in the laboratories. The study also suggests the guidance of laboratory testing of the isolates for ESBL production, so about to take proper precaution to prevent the spread of these resistant organisms to other patients and to begin effective treatment of patients. It is not possible to investigate the molecular methods routinely in the clinical laboratories of developing countries. But still steps will be taken to carry out to identify the exact genes involved in antibiotic resistance. The present study provides further authentication of the global sporadic of blaCTX-M and the relevance between K antigens, serum resistance, and ESBL production among the isolates of K. pneumoniae in our place. As there are no much study on virulence determined in respect to capsular antigen and genotypic detection of ESBLs, it also highlights the need for further study of their epidemiological surveillance.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Klebsiella spp. As nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589-603.
- [Google Scholar]
- Blood stream infections caused by extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae: Risk factors, molecular epidemiology, and clinical outcome. Antimicrob Agents Chemother. 2006;50:498-504.
- [Google Scholar]
- Third generation cephalosporin-resistance in K. pneumoniae isolates: An emerging threat. Int J Basic Clin Pharmacol. 2013;2:56-60.
- [Google Scholar]
- Mechanisms of bacterial resistance to antibiotics. Arch Intern Med. 1991;151:886-95.
- [Google Scholar]
- Detection of blaSHV $ bla CTX-M genes in ESBL producing K. pneumoniae isolated from Egyptian patients with suspected nosocomial infections. Egypt J Med Hum Genet. 2013;14:277-83.
- [Google Scholar]
- Occurrence of SHV, TEM and CTX-M β-lactamase genes among enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J Microbiol. 2015;8:e15620.
- [Google Scholar]
- Relation between blaTEM, blaSHV and blaCTX- M genes and acute urinary tract infections. J Acute Dis. 2016;5:71-6.
- [Google Scholar]
- Hypermucoviscosity as a virulence factor in experimental Klebsiella pneumoniae endophthalmitis. Invest Ophthalmol Vis Sci. 2008;49:4931-8.
- [Google Scholar]
- Serum resistance among Escherechia coli strain causing UTI in relation to O type and the carriage of hemolysin, colicin and antibiotic resistance determinants. Infect Immun. 1982;35:270-5.
- [Google Scholar]
- Virulence factors and drug resistance in Escherechia coli isolated from extra intestinal infections. Indian J Med Microbiol. 2007;25:369-73.
- [Google Scholar]
- Increased serum resistance in Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2004;48:3477-82.
- [Google Scholar]
- Bailey & Scott's Diagnostic Microbiology (1st ed). St. Louis: Mosby; 2002.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty First Informational Supplement M100-S21. Wayne, PA, USA: CLSI; 2011.
- [Google Scholar]
- Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867-78.
- [Google Scholar]
- Prevalence of bla TEM, bla SHV and bla CTX-M genes in clinical isolates of E.coli and Klebsiella pneumoniae from North East India. Indian J Pathol Microbiol. 2014;57:249-54.
- [Google Scholar]
- Molecular characterisation of extended spectrum beta lactamase producing strains from blood sample. Int J Pharm Pharm Sci. 2014;6:276-8.
- [Google Scholar]
- Multiplex PCR for bla(CTX-M) & bla(SHV) in the extended spectrum beta lactamase (ESBL) producing gram-negative isolates. Indian J Med Res. 2008;128:313-7.
- [Google Scholar]
- Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother. 2006;57:154-5.
- [Google Scholar]
- Phenotypic and Genotypic Characterization of Extended Spectrum Beta-Lactamases in Escherichia coli and Klebsiella pneumoniae Isolated across Karnataka (Doctoral thesis). Sri Devaraj Urs Academy of Higher Education and Research. Tamaka, Kolar, Karnataka, Microbiology Department. 2015
- [Google Scholar]
- Clinical and molecular analysis of Klebsiella pneumoniae causing liver abscess in China. J Mol Microbiol Biotechnol. 2016;26:245-51.
- [Google Scholar]
- A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697-705.
- [Google Scholar]
- An approach to uropathogenic Escherichia coli in urinary tract infections. J Lab Physicians. 2010;2:70-3.
- [Google Scholar]
- Variations in the prevalence of strains expressing an extended-spectrum beta-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific Region. Clin Infect Dis. 2001;32(Suppl 2):S94-103.
- [Google Scholar]
- Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci. 2015;22:90-101.
- [Google Scholar]
- Comment on: Occurrence, prevalence and genetic environment of CTX-M beta-lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother. 2007;59:799-800.
- [Google Scholar]
- Comparison of three dimensional test and double disc synergy test for detection of extended spectrum-lactamase (ESBL) producing gram negative bacteria. Pulse. 2013;6:12-619.
- [Google Scholar]
- Phenotypic and genotypic methods for detection of ESBL producing E. coli and K. pneumoniae isolates from ventilator associated pneumonia. J Clin Dign Res. 2013;7:1975-8.
- [Google Scholar]
- Extended-spectrum β-lactamases in gram negative bacteria. J Glob Infect Dis. 2010;2:263-74.
- [Google Scholar]
- Extended-spectrum beta-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect Dis. 2008;8:159-66.
- [Google Scholar]
- Prevalence of bla (CTX M) extended spectrum beta lactamase gene in Enterobacteriaceae from critical care patients. J Lab Physicians. 2011;3:80-3.
- [Google Scholar]
- Occurrence of the CTX-M, SHV and the TEM genes among the extended spectrum β-lactamase producing isolates of Enterobacteriaceae in a tertiary care hospital of North India. J Clin Diagn Res. 2013;7:642-5.
- [Google Scholar]
- Neonatal septicaemia caused by diverse clones of Klebsiella pneumoniae & Escherichia coli harbouring blaCTX-M-15. Indian J Med Res. 2013;137:791-9.
- [Google Scholar]
- Nosocomial outbreak of septicaemia in neonatal Intensive Care Unit due to extended spectrum β-lactamase producing Klebsiella pneumoniae showing multiple mechanisms of drug resistance. Indian J Med Microbiol. 2010;28:380-4.
- [Google Scholar]
- Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132:332-6.
- [Google Scholar]
- Faecal carriage of CTX-M-15-producing Klebsiella pneumoniae in patients with acute gastroenteritis. Indian J Med Res. 2009;129:599-602.
- [Google Scholar]
- Acquisition of extended-spectrum beta-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal Intensive Care Unit: Molecular epidemiology and analysis of risk factors. J Med Microbiol. 2010;59:948-54.
- [Google Scholar]
- Diversity of genotypes in CTX-M-producing Klebsiella pneumoniae isolated in different hospitals in Brazil. Braz J Infect Dis. 2011;15:420-5.
- [Google Scholar]
- CTX-M β-lactamase-producing Klebsiella pneumoniae in suburban New York city, New York, USA. Emerg Infect Dis. 2013;19:1803-10.
- [Google Scholar]
- Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013;13:466.
- [Google Scholar]
- Molecular characterization of CTX-M type extended spectrum beta lactamase producing E.coli isolated from humans and the environment. Indian J Med Microbiol. 2015;33(Suppl):73-9.
- [Google Scholar]
- Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47:3724-32.
- [Google Scholar]
- CTX-M: Changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165-74.
- [Google Scholar]
- Molecular characterization of extended-spectrum beta lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Dev Ctries. 2014;8:282-8.
- [Google Scholar]
- Multidrug resistance genes, including bla(KPC) and bla(CTX)-M-2, among Klebsiella pneumoniae isolated in Recife, Brazil. Rev Soc Bras Med Trop. 2012;45:572-8.
- [Google Scholar]
- Seroepidemiology of Klebsiella pneumoniae in an Australian tertiary hospital and its implications for vaccine development. J Clin Microbiol. 2006;44:102-7.
- [Google Scholar]





