Translate this page into:
Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniae
Address for correspondence: Dr. Poothakuzhiyil Remya, Department of Microbiology, Sri Ramachandra Medical College and Research Institute, Porur, Chennai - 600 116, Tamil Nadu, India. E-mail: remmoos@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Klebsiella pneumoniae causes both nosocomial and community-associated infections. Hypervirulent K. pneumoniae (hvKP), new variant of K. pneumoniae, can cause invasive infections in young healthy individuals as well as in the immunocompromised population. Hypervirulent strains frequently belong to capsular serotypes K1 or K2. Emergence of antimicrobial resistance in hvKP is a cause for concern.
AIM AND OBJECTIVE:
The present study was done to detect the K1 and K2 serotypes among clinical isolates of K. pneumoniae, spectrum of infections caused by them and presence of common beta-lactamases encoding genes in them.
MATERIALS AND METHODS:
A total of 370 isolates of K. pneumoniae, isolated from various clinical samples over a period of 1 year was included in this study. Antibiotic susceptibility testing to various classes of antimicrobials was done as per Clinical and Laboratory Standard Institute guidelines. The presence of K2A (specific to serotype K2), magA (specific to serotype K1), and rmpA genes was detected by multiplex polymerase chain reaction (PCR). Extended-spectrum beta-lactamases (TEM, SHV, and CTX-M), plasmid-mediated AmpCs (MOX, CIT, DHA, ACC, EBC, and FOX), and carbapenemase genes (IMP, VIM, NDM, KPC, and OXA-48) were also determined by PCR.
RESULTS:
Among the 370 isolates, 8 harbored K2A gene and one harbored magA. rmpA gene was detected in three isolates along with K1 or K2 serotypes. Seven K2A-positive isolates were resistant to one or more classes of antimicrobials. The studied ESBL genes were present in four isolates. Two isolates harbored carbapenemase genes (NDM-1, OXA-48) along with ESBLs.
CONCLUSION:
K2 serotype is more prevalent among hvKP isolates. They can harbor ESBLs and Carbapenemase genes. K1 serotype is rather uncommon in K. pneumoniae. Acquisition of multidrug-resistant genes by these strains adds to their virulence and limits the treatment options.
Keywords
Hypervirulent
K1 or K2 serotype
Klebsiella pneumoniae
molecular method
multidrug resistance
underlying diseases
Introduction
Klebsiella pneumoniae is a well-known human pathogen associated with both community-acquired and nosocomial infections. Classic K. pneumoniae (cKP) is the term used for the commonly recognized strains of K. pneumoniae, which cause bacteremia, pneumonia, urinary tract, and other infections. In 1986, hypervirulent K. pneumoniae (hvKP), a new variant of these bacteria, was first reported from Taiwan. Compared to cKP, hvKP strains produce hypermucoviscous colonies when grown on agar plates. They are capable of causing invasive and serious infections in immunocompromised as well as in healthy individuals.[12] Hypervirulent strains of K. pneumoniae are now circulating worldwide.[3]
There are a number of virulence factors that contribute to the pathogenicity of K. pneumoniae. They include hypermucoviscosity-specific capsular serotypes, especially K1 or K2, virulence gene FimH (fimbrial adhesion), rmpA (regulator of mucoid phenotype), uge (UDP-glucose 4-epimerse), kfu (an iron uptake system), and alls (allantoin metabolism).[45]
K. pneumoniae is known to synthesize capsules, which are essential for its virulence. At least, 78 capsular serotypes exists in this bacteria. Among hvKP, eight capsular serotypes occur; of them K1 and K2 are the most virulent that have been described to date. The capsules of K1 and K2 serotype essentially protect the bacteria from phagocytosis.[46]
K1 or K2 serotypes are more prevalent in invasive infections.[7] Serotype-specific gene for K1 is magA (mucoviscosity associated gene A), and for K2 serotype, it is K2A (K2 capsule-associated gene A). magA gene was first described by Fang et al. in 2004 and Chuang et al. found that it is restricted to the gene cluster of capsular serotype K1. Similarly, K2A gene, which corresponds to magA in K1 serotype, is highly specific to the cps gene cluster of capsular serotype K2. Detection of both magA and K2A is a rapid and accurate method for the detection of K1 and K2 serotypes of K. pneumoniae.[89] rmpA (regulator of mucoid phenotype A) gene is a plasmid-mediated regulator of capsular polysaccharide synthesis. It is also associated with hypermucoviscosity phenotype and invasive disease.[28]
The past hypervirulent strains of K. pneumoniae were susceptible to all antimicrobials except ampicillin.[6] However, the first report from China in 2014 described that the clonal complexes of hvKP can be MDR.[3]
This study was conducted to detect the K1 and K2 serotypes among clinical isolates of K. pneumoniae, the spectrum of infections caused by them and the presence of common beta-lactamases encoding genes in them.
Materials and Methods
Bacterial isolates
The study was conducted in a University teaching hospital in South India. A total number of 370 clinically significant, consecutive, nonduplicate isolates of K. pneumoniae, collected from September 2014 to August 2015 were included in this study.
The source of isolates were urine (n = 170), exudative specimens (n = 132), respiratory secretions such as bronchial wash, endotracheal aspirate, pleural fluid (n = 38), and blood (n = 30).
The isolates were identified up to species level by automated system (VITEK2 GN-card; BioMerieux, Brussels, Belgium) and/or standard biochemical tests.[10]
The clinical significance of all isolates was determined by significant colony count, presence of bacteria in gram stain and correlating clinical history.
Phenotypic detection of hyperviscosity
Hyperviscosity of the isolates was tested by evaluating the formation of a mucoviscous string of >5 mm, when using an inoculation loop to stretch a colony grown on an agar plate.[1]
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was done by Kirby–Bauer disc diffusion method for different classes of antibiotics such as cefotaxime (30 μg), ceftazidime (30 μg), cefoxitin (30 μg), amikacin (30 μg), and ciprofloxacin (5 μg), piperacillin/tazobactam (100 μg/10 μg), and imipenem (10 μg) (Himedia laboratories, Mumbai, Maharashtra, India) as per Clinical and Laboratory Standard Institute guidelines.[11] ATCC Escherichia coli 25,922 was used as control strain.
The clinical history was sought for all the K1 and K2 serotypes from the medical records.
Molecular methods
Multiplex polymerase chain reaction for virulence-associated genes
Bacterial DNA was extracted by boiling method. Genomic DNA was used as the template and multiplex polymerase chain reaction (PCR) was carried out to amplify magA (specific for K1serotype), K2A (specific for K2 serotype), and rmpA genes as per earlier described methods.[5] PCR was performed with a final volume of 25 μl. Each reaction contained 10 pmol of each primer (Sigma-Aldrich, India),10mM of dNTP mixture (Takara, India), 5U Taq polymerase (Takara, India) in 2.5μl of 10X Taq polymerase buffer (Mg2+plus). A volume of 2 μl of template DNA was added to 23 μl of the master mixture. Negative controls were PCR mixture with water (instead of template DNA) and were included in every PCR run.
The PCR conditions were as follows: initial activation at 95°C for 15 min, followed by 30 cycles at 94°C for 30 s, annealing at 60°C for 90 s, extension at 72°C for 60 s, and the final extension was at 72°C for 10 min. The amplicons were separated at 100 V in a 1.5% agarose gel containing ethidium bromide.
The primers used are summarized in Table 1. Previously, characterized strains were used as positive controls.

Detection of beta-lactamase encoding genes by polymerase chain reaction
For the study isolates, PCR was performed to detect the presence of common lactamase genes namely, ESBLs (TEM, SHV, and CTX-M), plasmid-mediated AmpCs (MOX, CIT, DHA, ACC, EBC, and FOX), and Carbapenemase genes (IMP, VIM, NDM, and KPC and OXA-48). The primers used for beta-lactamase genes were selected based on the previous studies.[121314]
DNA sequencing
PCR positive amplicons were purified and sequenced by big dye 3.1 cycle sequencing kit using the Sanger AB13730XL DNA analyzing instrument (Sci genome, India). The nucleotide sequences analyzed were compared with the sequence available at the National Centre for Biotechnology information website (www.ncbi.nIm.nih.gov).
The sequences were submitted to the genbank with the following accession numbers MF188922 (K2A), MF188924 (magA), and MF188921 (rmpA).
Results
Of the 370 isolates 9 (2.4%) carried one or more studied virulence genes, and all the nine isolates were positive for the string test. Six isolates of them carried only K2A. One isolate harbored magA along with rmpA. Genes for K2A and rmpA coexisted in two isolates. Gel picture of the amplified genes was showed in Figure 1.
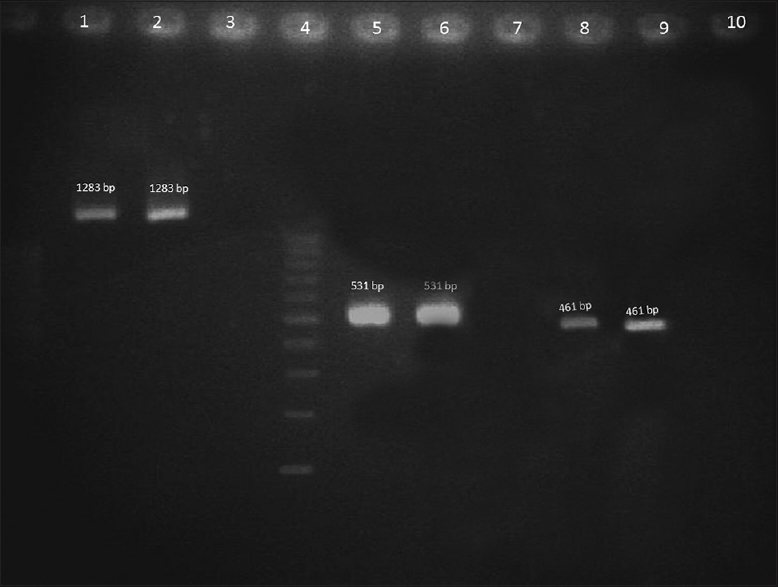
- Gel picture of the amplified genes. Lane 1-magA test strain, Lane 2 – magA-positive control, Lane 3 - Negative control, Lane 4 – 100 bp Ladder, Lane 5-K2A test strain, Lane 6 - K2A positive control, Lane 7 - Negative control, Lane 8-rmpA test strain, Lane 9 – rmpA-positive control, and Lane 10 - Negative control
Of the K1, K2 serotypes serotype positive isolates, two were susceptible to all classes of antibiotics tested while the others (n = 7) exhibited resistance to one or more classes of antibiotics. The resistance exhibited to various classes of antibiotics were as follows cefotaxime 100% (7/7), ceftazidime 100% (7/7), cefoxitin 57% (4/7), amikacin 42.8% (3/7), ciprofloxacin 57% (4/7), piperacillin/tazobactam 71.4% (5/7), and imipenem 28.5%(2/7).
The occurrence of the studied beta-lactamase genes along with virulence is shown in Table 2. Notably, plasmid-mediated AmpC genes were not detected in any of the K1 or K2 serotypes.

All three studied ESBL genes, TEM+SHV+CTX-M together were detected along with K2A gene in 4 isolates of K. pneumoniae, and one harbored two ESBL genes (SHV+CTX-M). SHV alone was found in one K2A-positive isolate. Only one magA-positive isolate in this study carried blaSHV.
Of the eight K2A positives, two isolates harbored NDM and OXA-48 along with all three ESBL genes.
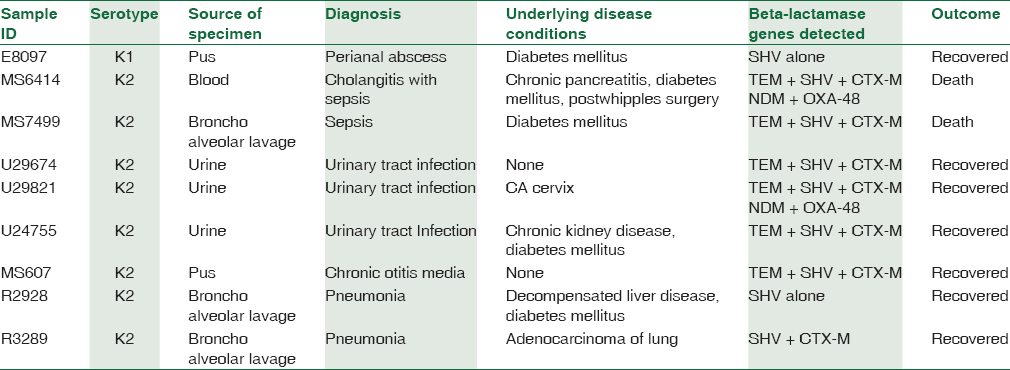
Clinical profile of the patients harboring K1, K2 serotypes and the beta-lactamse genes detected in them are shown in Table 3.
Discussion
K. pneumoniae is a common nosocomial and a potential community-acquired pathogen which can cause infections in all age groups, especially in the immunocompromised. The emergence of hypervirulent strains broadens the occurrence of infection among healthy and immunocompetent individuals also.[1516] Several virulence factors are identified in K. pneumoniae including capsules, lipopolysaccharides, siderophores, and fimbriae. Of these, capsule is the most studied virulence factor. Among the capsular serotypes identified to date, K1 and K2 serotypes are mostly reported in invasive diseases and known to be hypervirulent or hypermucoviscous.[17]
In this study, 2.16% of the total isolates only harboured K2A gene (specific to K2 serotype), and only one carried magA gene (specific to K1 serotype). In the present study, K2 serotype is the most predominant among hyperviscous K. pneumoniae. This is in contrast to observations made in other countries. K1 is the most predominant serotype in a study from east China (68.9%) followed by K2 serotype (20%). They were isolated from liver abscess patients.[2] In Asia, K1 is the main serotype followed by K2.[18] A study from Germany in 2014 reported one K1 and one K2 serotype in liver abscess patients.[18] In Taiwan, K1 serotype was detected in 63.4% of liver abscess, and K2 in 14.2%.[19] A publication from Japan in 2016, described K1 serotype in one and K2 serotype in two liver abscess patients.[20] The presence of K1 or K2 serotypes are also being reported from North America, Europe, Australia, Africa, and the Middle East.[6] Although K1/K2 serotypes are frequently associated with liver abscess, they have been isolated from extrahepatic abscess also.[241821]
In this study, three isolates coharbored rmpA gene along with K1and K2 serotypes. Association of rmpA gene was found in 55% (16/29) of isolates elsewhere.[1] Occurrence of rmpA gene along with K2 serotypes has been reported from France and Japan also.[320]
In the past, hvKP were susceptible to all classes of antibiotics, except ampicillin.[6] In this study, only two isolates among the nine K1 or K2 serotypes were susceptible to all classes of antibiotics tested. The seven K2A positive isolates were resistant to one more classes of antimicrobials tested. A study from France reported resistance to third-generation cephalosporins and to all aminoglycosides except Amikacin among K2 serotypes.[3] Another study by Zhang et al. reported resistance to all classes of antibiotics including carbapenems among K1 and K2 serotypes.[22]
Antibiotic resistance is commonly associated with cKP, and till 2014, it was believed that hvKP is independent of multidrug resistance.[2223] Only two isolates with SHV-1 was detected from K1 and K2 serotypes in this study. This SHV-1 is a chromosomal gene inherently found in all K. pneumoniae. The presence of ESBL genes, such as TEM, SHV, and CTX-M, were detected in 4 (4/8) of K2 serotypes. Surgers L et al (2016). observed CTX-M3-producing strain of K2 hvKP in a patient with liver abscess (n = 3).[3] Another study from China, detected 17% (5/29) of hvKP producing ESBLs.[1]
Carbapenemase genes were detected in two K2 serotypes in this study. In both the isolate, NDM 1 and OXA 48 were present along with ESBL (TEM, SHVand CTX M) genes. To the best of our knowledge, no study has yet reported the presence of NDM and OXA-48 together in a K2 serotype. The presence of NDM-1 in K1 serotype was reported by Fabrice Compain et al. in a recent study.[24] Mei et al. in 2017 described the presence of NDM-5 in K2 serotype.[25] Another study from China highlighted the occurrence of KPC-2 along with K1 hvKP.[22]
In this study, K1 serotype was detected in one patient with perianal abscess. In Korea, 4.9% of K. pneumoniae isolated from stool specimens were of K1 serotype.[26] Liu et al. opined that K. pneumoniae is one of the major causative agents for perianal abscess in diabetic patients.[27]
In the present study, fatal outcome was observed in two patients with K2 K. pneumoniae. One of them had sepsis due to cholangitis caused by K2 serotype harboring TEM, SHV, CTX-M, NDM, and OXA 48. The other patient also had sepsis associated with pneumonia caused by K2 serotype harboring TEM, SHV, and CTX-M. Regard to underlying or comorbid condition, two patients in this study had no identifiable underlying disease. Five had diabetes mellitus. This is one of the well-recognized risk factor for hvKP infections. Malignancy, chronic kidney disease, and chronic liver disease were the other risk factors identified in the study patients.
Conclusion
Acquisition of MDR genes by hvKP strains is worrisome and should be closely monitored. Management of infections caused by these will become extremely challenging because the outcome will be worse with inappropriate treatment. A combination of multidrug resistance and hypervirulence will lead to poorer outcomes. Detection of genes determining the hyperviscosity is therefore important for anticipating severe infections with invasive spread. Molecular typing of the hvKP can help to determine clonal relatedness of the strains and give insights with their epidemiology.
Financial support and sponsorship
This study was financially supported by the Founder Chancellor Ramasamy Udayar Fellowship provided by Sri Ramachandra Medical College and Research Institute.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
All the authors are grateful for the financial assistance provided by Sri Ramachandra Medical College and Research Institute as Founder Chancellor Fellowship.
References
- Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58:225-32.
- [Google Scholar]
- Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis. 2015;15:161.
- [Google Scholar]
- ESBL-producing strain of hypervirulent Klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22:1687-8.
- [Google Scholar]
- Clinical spectrum and molecular characteristics of Klebsiella pneumoniae causing community-acquired extrahepatic abscess. J Microbiol Immunol Infect. 2008;41:311-7.
- [Google Scholar]
- Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52:4377-80.
- [Google Scholar]
- Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence. 2013;4:107-18.
- [Google Scholar]
- Impaired phagocytosis of capsular serotypes K1 or K2 Klebsiella pneumoniae in type 2 diabetes mellitus patients with poor glycemic control. J Clin Endocrinol Metab. 2006;91:3084-7.
- [Google Scholar]
- Multiplex-PCR assay for identification of Klebsiella pneumoniae. Int J Pharm Sci Rev Res. 2014;26:112-7.
- [Google Scholar]
- Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis. 2007;195:1235-6.
- [Google Scholar]
- Test for identification of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A, eds. Mackie and McCartney Practical Medical Microbiology (14th ed). Edinburgh: Churchill Livingstone; 1996. p. :131-49.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. In: 25th Informational Supplement Document M100-S25. Wayne: Clinical and Laboratory Standards; 2015.
- [Google Scholar]
- Detection of extended spectrum beta-lactamases-producing isolates and effect of AmpC overlapping. J Infect Dev Ctries. 2013;7:618-29.
- [Google Scholar]
- Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153-62.
- [Google Scholar]
- Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119-23.
- [Google Scholar]
- Klebsiella pneumoniae bacteremia: Community-acquired vs. Nosocomial infections. Chang Gung Med J. 2001;24:688-96.
- [Google Scholar]
- Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629-61.
- [Google Scholar]
- The contribution of capsule polysaccharide genes to virulence of Klebsiella pneumoniae. Virulence. 2017;8:485-6.
- [Google Scholar]
- Klebsiella pneumoniae-induced liver abscesses, Germany. Emerg Infect Dis. 2014;20:1939-40.
- [Google Scholar]
- A global emerging disease of Klebsiella pneumoniae liver abscess: Is serotype K1 an important factor for complicated endophthalmitis? Gut. 2002;50:420-4.
- [Google Scholar]
- Invasive liver abscess syndrome caused by Klebsiella pneumoniae with definite K2 serotyping in Japan: A case report. Surg Case Rep. 2016;2:72.
- [Google Scholar]
- K2 serotype Klebsiella pneumoniae causing a liver abscess associated with infective endocarditis. J Clin Microbiol. 2010;48:639-41.
- [Google Scholar]
- Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60:709-11.
- [Google Scholar]
- Invasive Klebsiella pneumoniae infections, California, USA. Emerg Infect Dis. 2010;16:1490-1.
- [Google Scholar]
- Primary osteomyelitis caused by an NDM-1-producing K. Pneumoniae strain of the highly virulent sequence type 23. Emerg Microbes Infect. 2017;6:e57.
- [Google Scholar]
- Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM-5 in China. Front Microbiol. 2017;8:335.
- [Google Scholar]
- Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis. 2012;31:481-6.
- [Google Scholar]
- Clinical and microbiological analysis of adult perianal abscess. J Microbiol Immunol Infect. 2011;44:204-8.
- [Google Scholar]





