Translate this page into:
Phenotypic and genotypic characterization of multidrug-resistant isolates from patients with catheter-associated urinary tract infection in a tertiary care hospital
Address for correspondence: Dr. Nirupa Soundararajan, Department of Microbiology, Chettinad Hospital and Research Institute, Chettinad Academy of Research and Education, Kelambakkam - 603 103, Tamil Nadu, India. E-mail: drnirups@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
INTRODUCTION:
Urinary tract infections (UTIs) remain as the most common infection. Catheter-associated (CA) UTI can lead to bacteremia and thereby is the leading cause of morbidity and mortality in hospitalized patients in our country.
AIMS AND OBJECTIVES:
This study aims to check the prevalence of CAUTI and study the phenotypic and genotypic characters of the multidrug-resistant organisms in a tertiary care hospital, with special reference to NDM-1 and OXA-23.
MATERIALS AND METHODS:
A total of 231 urine samples from patients with CA-UTI in different wards in a tertiary care hospital over a period of 3 months between June and August 2018 were collected and processed following the standard protocol. Antibiotic susceptibility tests were performed by disk-diffusion method. Modified Hodge test (MHT) was done to isolate carbapenem-resistant isolates, and polymerase chain reaction was done to detect NDM-1 and OXA-23.
RESULTS:
Out of 231 samples, 101 samples yielded significant growth. These 38 samples were Gram-negative bacilli which were resistant to carbapenems. Out of the 38 which showed carbapenem resistance, 23 were MHT positive. Out of the 23 MHT-positive isolates, 8 (21.05%) were positive for NDM-1 gene and only 1 (2.6%) was positive for the OXA-23 gene.
CONCLUSION:
This study has shown that carbapenem-resistant isolates from all the CA urinary tract-infected patients were 52.77% and most of them were Klebsiella. About 21% of them harbored the NDM-1 gene whereas only 2% had the OXA-23 gene. There has been an alarming increase in the spread of carbapenem resistance.
Keywords
Carbapenem resistance
catheter-associated urinary tract infection
hospital-acquired infection
multidrug-resistant bacteria
NDM-1 gene
OXA-23 gene
urinary catheter
Introduction
Urinary tract infections (UTIs) are the most common infections occurring in people. In critically ill patients, catheter-associated (CA) UTIs can lead to bacteremia, and they are the leading cause of morbidity and mortality in approximately 10% of hospitalized patients.[1] CA-UTI accounts for over 80% of infections in catheterized patients admitted in intensive care units (ICUs) during their hospital stay.[2] The important predisposing factors commonly associated with CAUTIs are conditions such as diabetes, immunosuppression, renal insufficiency, and urinary incontinence commonly among neurologic and orthopedic patients.[12] Other factors for CAUTI could be patients staying on catheter for a prolonged duration of time, female and elderly patients, patients with severe illness, catheterization performed under nonsterile conditions, and catheter insertion by undertrained professionals.[34] The important factor that help to reduce CAUTIs is catheterizing the patient only when absolutely necessary and by shortening the duration time of indwelling catheters. The incidence of multidrug resistance in organisms is increasing due to dissemination of resistance determinant genes mediated by transposons, plasmids, and gene cassettes in integrons.[3] Beta-lactam NDM-1 is a novel metallo-beta-lactamase with divalent Zn ions at its active site and has the ability to confer resistance to almost all the β-lactams. The NDM-1 gene is normally carried on a variety of plasmids along with other resistance factors. The bacteria also show broad antibiotic resistance to all classes except colistin. OXA-23 may be chromosomally and plasmid mediated and encodes for serine carbapenemases widely present among Acinetobacter species and is responsible for its extreme drug resistance. Very rarely, OXA-23 group is seen in Enterobacteriaceae. Resistance to carbapenems due to carbapenemase production poses serious challenges in the treatment of such infections with pan-resistant phenotypes.[5] Spreading of NDM-1 gene in a hospital may be a complex event involving several modes of spread.[6] This study aims to calculate the prevalence of CAUTI, the common pathogens involved, to phenotypically and genotypically characterize the multidrug-resistant isolates from CAUTI patients, and to determine the prevalence of NDM-1 and OXA-23 genes involved in carbapenem resistance.
Materials and Methods
Samples were collected after obtaining informed consent from the patients or their attendants. Catheterized urine sample was collected aseptically following clamping and using a sterile collection container. The urine was collected through the catheter port. After cleaning the catheter with 70% isopropyl alcohol, it was clamped. After 30 min with the help of a sterile syringe, 1–2 ml of urine was aspirated and collected in a sterile container. The clamp was removed. Adult patients who were catheterized belonging to both sexes and admitted as inpatients in various wards such as ICU, medical wards, surgical wards, obstetric and gynecology wards, and orthopedic wards were included in this study. Samples were collected only from patients in whom the catheter has been in situ for a minimum of 48 h duration. Patients with preexisting renal disease were excluded from the study.
The urine sample collected from catheter was transported to the clinical microbiology laboratory without delay, within 1–2 h of sample collection, as urine is an excellent culture medium and contaminating bacteria might multiply and reach significant numbers.
Gram's stain of a smear made from urine was performed, and the presence of pus cells along with Gram-negative bacilli, Gram-positive cocci, and Gram-positive budding yeast cells was noted. The clinical significance of growth in urine cultures was estimated by the colony count of the organism per milliliter of urine. The urine was cultured on cysteine–lactose–electrolyte-deficient agar. This plate was incubated at 37°C for 18–24 h, and colony morphology was observed and noted the next day. As indwelling catheter specimens are collected through the urethra, the urine is likely to be contaminated by normal urethral flora, and colony counts are necessary for interpretation of culture. Samples found to have mixed growth of organisms were rejected, and a repeat sample was collected. Samples having >105 colony-forming units per milliliter were processed further to identify the pathogen. For Gram-positive cocci and yeast, counts of 104–105 CFU/ml were considered significant. In general, counts <103 CFU/ml were considered as contamination. In Gram's stain, the presence of pus cells and bacteria in every oil immersion field/20 fields indicate infection. Ninety-five percent of all UTIs were caused by a single type of organism. The routine biochemical tests for organism identifications such as indole test, triple sugar iron test, citrate utilization test, urease production test, mannitol-motility medium, and phenyl deaminase test were performed.
Antimicrobial susceptibility testing (Kirby–Bauer disk-diffusion method)
Antibiotic susceptibility test was performed by Kirby–Bauer disk-diffusion methods on Mueller–Hinton agar using 0.5 McFarland standard turbidity of bacterial inoculum according to the CLSI guidelines. Antibiotic discs used were ampicillin (10 μg), cefazolin (30 μg), cefuroxime (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), piperacillin/tazobactam (100/10 μg), gentamicin (10 μg), amikacin (30 μg), nitrofurantoin (300 μg), norfloxacin (10 μg), ciprofloxacin (5 μg), cotrimoxazole (1.25/23.75 μg), imipenem (10 μg), and meropenem (10 μg).
Phenotypic methods for identifying carbapenem resistance
Those isolates which were resistant to imipenem and meropenem in addition to beta-lactams were subjected to the modified Hodge test (MHT). This test detects carbapenemase production in isolates of Enterobacteriaceae but does not differentiate between serine and metallo-beta-lactamases. This test was carried out as per the CLSI guidelines 2017. A 0.5 McFarland standard suspension of Escherichia coli ATCC 25922 was prepared and inoculated in a Mueller–Hinton agar plate as for the routine disk-diffusion procedure. The plate was allowed to dry for 3–10 min. Place the meropenem disk on the plate in the center. Using a 10 μl loop or swab, 3–5 colonies of test organism grown overnight on a blood agar plate were inoculated in a straight line out from the edge of the disk. The streak should be at least 20–25 mm in length. Six organisms were tested on a plate and incubated at 35°C ± 2°C in ambient air for 16–20 h. Indentation of growth (clover-shaped zone of inhibition) of ATCC E. coli close to streak of isolate-producing carbapenemase is seen and interpreted as positive MHT. Positive and negative controls used were Klebsiella pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC BAA-1706, respectively.
Genotypic method
Polymerase chain reaction (PCR) for the detection of NDM-1 and OXA-23 genes was carried out for pathogens showing positive MHT.
DNA extraction
Bacterial DNA Mini Spin purification kit was used for DNA extraction (kit contains lysozyme, lysozyme digestion buffer, proteinase K, binding buffer, wash buffer-1, wash buffer-2, spin columns with collection tube, and elution buffer.
The primer used for OXA-23 (453bp) was:
OXA-23-F AGTATTGGGGCTTGTGCT
OXA-23-R AACTTCCGTGCCTATTTG
The primer used for NDM-1 (214bp) was:
NDM-1-F GCAGCACACTTCCTATCTCG
NDM-1-R GTCCATACCGCCCATCTTGT
Polymerase chain reaction amplification procedure and gel documentation
One microliter of the extracted DNA was added to the PCR mixture. Initial denaturation was done at 94°C for 3 min followed by primer annealing at 55°C for 1 min. Heat-stable extension was done at 72°C for 1 min 30 s. The total number of cycles carried out was 32. The final extension was done at 72°C for 7 min.
HELINI 2X ReddyMix PCR Master Mix, agarose gel electrophoresis consumables, and NDM-1 and OXA-23 primers were procured from HELINI Biomolecules, Chennai, India. The process of electrophoresis was monitored by observing the migration of a visible dye (tracking dye) through the gel (bromophenol blue) moving at the same speed as that of the double-stranded DNA. Ethidium bromide dye was added to agarose gel, and isolation of bands was determined by the examination of the gel under the ultraviolet transilluminator. Expected product size was 214 bp for NDM-1 and 453 bp for OXA-23. Appropriate controls were used.
Results
A total number of 231 urine samples were collected from catheterized patients over a period of 3 months between June and August 2018 out of which 133 (57.6%) were males and 98 were females (42.4%). Seventy percent of these samples were from the ICU, and the rest were from other wards such as medicine, surgery, obstetrics and gynecology, and orthopedics. Of these, 101 samples (43.7%) yielded growth while the rest showed no/insignificant bacteriuria. Table 1 shows the organisms isolated in culture. Klebsiella was the most common organism to be isolated. Tables 2 and 3 show the antibiotic susceptibility pattern of all the Gram-negative bacilli and Gram-positive cocci isolated from these samples.
| Growth on culture | Number of isolates (%) |
|---|---|
| K. pneumoniae | 25 (26.3) |
| E. coli | 23 (24.2) |
| Pseudomonas spp. | 15 (15.8) |
| P. vulgaris | 2 (2.1) |
| Providencia spp. | 2 (2.1) |
| Acinetobacter spp. | 4 (4.2) |
| M. morganii | 1 (1.5) |
| Enterococcus spp. | 9 (9.5) |
| Candida spp. | 20 (21.05) |
M. morganii=Morganella morganii, K. pneumoniae=Klebsiella pneumoniae, P. vulgaris=Proteus vulgaris, E. coli=Escherichia coli
| Organisms | Resistance pattern (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CZ | CXM | CTX | CPM | PIT | GEN | AK | NIT | NX | CIP | COT | IMP | MRP | |
| Klebsiella spp. | 100 | 100 | 100 | 100 | 100 | 100 | 92 | 100 | 88 | 100 | 100 | 100 | 76 | 76 |
| E. coli | 52.8 | 39 | 39 | 34.7 | 34.7 | 21.7 | 17.3 | 8.7 | 13 | 47 | 35 | 22 | 34.7 | 34.7 |
| Acinetobacter spp. | 50 | 50 | 50 | 50 | 50 | 25 | 50 | 50 | 50 | 50 | 50 | 50 | 75 | 75 |
| Pseudomonas spp. | - | 20 | - | - | 13.3 | 20 | 13.3 | 13.3 | - | - | 13.3 | - | 47 | 47 |
| M. morganii | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
AMP: Ampicillin, CZ: Cefazolin, CXM: Cefuroxime, CTX: Cefotaxime, CPM: Cefepime, PIT: Piperacillin tazobactam, GEN: Gentamicin, AK: Amikacin, NIT: Nitrofurantoin, NX: Norfloxacin, CIP: Ciprofloxacin, COT: Cotrimoxazole, IMP: Imipenem, MRP: Meropenem, M. morganii=Morganella morganii, E. coli=Escherichia coli
| Organisms | Resistance pattern (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | P | AMP | CIP | HLG | TEI | COT | VA | LZ | NIT | NX | |
| Enterococcus | 9 | 11 | 11 | 44 | 33 | 33 | 33 | 0 | 0 | 11 | 33 |
AMP: Ampicillin, NIT: Nitrofurantoin, NX: Norfloxacin, CIP: Ciprofloxacin, COT: Cotrimoxazole, CIP: Ciprofloxacin, HLG: High level Gentamicin, TEI: Teicoplanin, VA: Vancomycin, LZ: Linezolid, P: Penicillin
A total of 38 Gram-negative bacilli were resistant to carbapenems, of which 29 (76%) were from the ICU, 6 (16%) were from surgical wards, and 3 (8%) were from the medical wards. Of the 38 carbapenem-resistant Gram-negative bacilli, 23 were MHT positive and 15 were negative. Table 4 gives the distribution of organisms which were MHT positive.
| Organisms | Total number of organisms (GNB) | Imipenem resistant | Meropenem resistant | Modified Hodge test positive |
|---|---|---|---|---|
| Klebsiella spp. | 25 | 19 | 19 | 13 |
| E. coli | 23 | 11 | 11 | 7 |
| Pseudomonas spp. | 15 | 5 | 5 | 1 |
| Acinetobacter spp. | 4 | 2 | 2 | 1 |
| M. morganii | 1 | 1 | 1 | 1 |
| Proteus spp. | 2 | - | - | - |
| Providencia | 2 | - | - | - |
| Total | 72 | 38 | 38 | 23 |
GNB=Gram negative bacilli, M. morganii=Morganella morganii, E. coli=Escherichia coli
Table 5 shows the PCR analysis of NDM-1 gene, and Table 6 shows the PCR analysis of OXA-23 gene. Figures 1 and 2 show gel documentation of the same.
| Organism which is positive for NDM-1 | NDM-1 positive |
|---|---|
| Klebsiella spp. | 4 |
| E. coli | 2 |
| Acinetobacter spp. | 1 |
| M. morganii | 1 |
| Total | 8 |
NDM=New Delhi metallo-beta-lactamase, M. morganii=Morganella morganii, E. coli=Escherichia coli
| Organism which is positive for OXA-23 | OXA-23 positive |
|---|---|
| Klebsiella spp. | 0 |
| E. coli | 0 |
| Acinetobacter spp. | 1 |
| M. morganii | 0 |
| Total | 1 |
M. morganii=Morganella morganii, E. coli=Escherichia coli

- Polymerase chain reaction of NDM-1 was positive in 1, 9, and 10 and the band was 214 bp
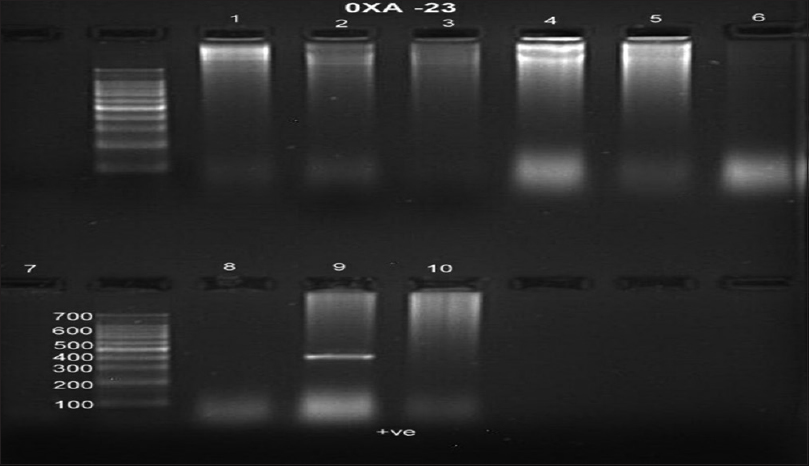
- Polymerase chain reaction of OXA-23 was positive in 9 and the band was more than 453 bp
In this study, 101 organisms were isolated, of which 72 were Gram-negative bacilli. Out of this, 38 showed carbapenem resistance, in that 23 were positive for MHTs and 8 (21.05%) were positive for NDM-1 gene. Of the 38 organisms tested for OXA-23 gene, only 1 (2.6%) was positive for the OXA gene.
Discussion
Of 231 samples collected, 95 samples were culture positive, of which 72 isolates were Gram-negative bacilli. This shows that the overall incidence of infection is 41%. Age-wise distribution revealed that maximum incidence was seen in those above 60 years of age. Bagchi et al.[7] in their study period of 2 years followed up 220 patients admitted at various medical, surgical wards and ICUs with respect to clinical features, risk factors, microbiological profile of CAUTI, and antibiotic susceptibility and found that 64 cases developed CAUTI and the incidence rate was 29.09%. This incidence is lower than that of the present study. Kazi et al.[8] studied 1380 catheterized patients, out of which 34 developed CAUTI. He demonstrated that if infection control practices are in place and monitored meticulously, it is possible to keep the incidence rates within the benchmark set by the individual hospital. In the present study, samples collected from ICUs were 70.1% and wards were 29.9%. Urine samples collected from ICUs had increased incidence of Candida spp. while those from other wards had E. coli in high incidence. However, the predominant uropathogen isolated from samples in our study was Klebsiella species, and E. coli was the second most common CA uropathogen after Klebsiella. Among the most common pathogens isolated in the ICUs, Candida spp. and Klebsiella spp. are most frequently encountered. In another study by Hanumantha and Pilli,[9] the predominant uropathogen was Citrobacter species, which was not isolated in this study. In a study by Vinod et al.[10] in Vellore, E. coli followed by Klebsiella sp were the common isolates in CAUTI. Therefore, it is inferred that the predominant pathogen varies from region to region depending on the local microbiological environment. In this study, the antibiotic susceptibility test result for the various organisms shows that all the Klebsiella isolates, Morganella morganii, and Proteus species and 50% of E. coli, Acinetobacter species, and most of the Pseudomonas isolates were resistant to the usual first and second-line antibiotics recommended for UTI, such as beta-lactams, aminoglycosides, and fluoroquinolones. In addition, increased resistance was seen for carbapenems such as imipenem and meropenem which leaves the clinician with no choice except for polymyxins. Organisms that produce carbapenemase enzyme are multidrug resistant. Increasing resistance to carbapenems that are most often the last line of therapy is being noted in hospital. The resistance to carbapenems may be related to either the decreased membrane permeability with overexpression of beta-lactamase or to expression of carbapenemases. In a study conducted by Agarwal et al.[11] in a tertiary care hospital in Northern India, 9.4% of E. coli isolates were positive for carbapenemase production and 7.2% isolates showed the presence of NDM-1 gene. In a study conducted in North East India by Bora et al.,[12]5% of E.coli isolates harbored the NDM-1 gene. Brennan et al.[13] reported carbapenem-resistant Enterobacteriaceae (CRE) in 61% of isolates from urine cultures, and a urinary catheter was present in 48% of these patients. Several studies carried out in different parts of India also have yielded similar results. Table 7 gives a comparative analysis of such studies which clearly shows that there has been an increase in drug resistance and prevalence of NDM-1 gene. In this study, out of the 38 CRE, 8 isolates were positive for the NDM-1 gene. Of these 8 isolates, 4 were Klebsiella, 2 were E. coli, Acinetobacter and Morganella one each, and one sample was positive for OXA-23. New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii is a major concern. The presence of NDM and its variants were studied in P. aeruginosa and A. baumannii at a tertiary hospital in North India by Rahman et al.,[14] and it has been found that 29.23% P. aeruginosa and 18.8% A. baumannii isolates were resistant to carbapenems and all of them were blaNDM positive. The single isolate which was positive for OXA-23 was Acinetobacter species. According to Khurana et al.,[15] the carbapenem resistance was higher in North than in South India. Gram-negative Bacilli blaOXA-1 was the most common carbapenemase gene among the South India strains, while blaNDM-1 followed by OXA-1 were the two most prevalent carbapenemase genes in the North Indian isolates.
| This study | Other studies | ||
|---|---|---|---|
| Commonest bacteria | Klebsiella species, E. coli | Hanumantha and Pilli, Vishakapatnam[9] | Citrobacter spp., Pseudomonas aeruginosa |
| Vinoth et al., Vellore[10] | E. coli, Klebsiella spp. | ||
| Antibiotic resistance | 53% carbapenemase producers | Agarwal et al., Uttar Pradesh[11] | 12.2% carbapenemase producers |
| NDM-1 gene prevalence | 11% | Agarwal et al., Uttar Pradesh[11] | 7.2% |
| Bora et al., North east India[12] | 5% |
E. coli=Escherichia coli, NDM=New Delhi metallo-beta-lactamase
Conclusion
This study has shown that carbapenem-resistant isolates from all the CA urinary tract-infected patients were 52.77% and most of them were Klebsiella. About 21% of them harbored the NDM-1 gene while only 2% had the OXA-23 gene. As all these genes confer multidrug resistance or pandrug resistance, it is very difficult to treat and adds to the morbidity and mortality rates. OXA-23 group is very notorious in spreading drug resistance among the superbugs Acinetobacter spp. So far it is not common among other Gram-negative bacteria, but as it is also plasmid mediated, we can soon expect it to spread and cause pandrug resistance among other organisms which are commonly isolated. There has been an alarming increase in the spread of carbapenem resistance which makes treatment of the patients very difficult, as most of the catheterized patients are in the ICUs and have several other associated comorbidities. To reduce the incidence of CA UTIs, it will be better if hospital infection control practices are in place.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Multimodal supervision programme to reduce catheter associated urinary tract infections and its analysis to enable focus on labour and cost effective infection control measures in a tertiary care hospital in India. J Clin Diagn Res. 2012;6:1372-6.
- [Google Scholar]
- Strategies for preventing catheterassociated urinary tract infections. Int J Prev Med. 2018;9:50.
- [Google Scholar]
- Emergence of carbapenem nonsusceptible multidrug resistant Acinetobacter baumannii strains of clonal complexes 103(B) and 92(B) harboring OXA-type carbapenemases and metallo-β-lactamases in Southern India. Microbiol Immunol. 2015;59:277-84.
- [Google Scholar]
- Urinary tract infections in the critical care unit: A brief review. Indian J Crit Care Med. 2013;17:370-4.
- [Google Scholar]
- Emergence of NDM – 1 in the clinical isolates of Pseudomonas aeruginosa in India. J Clin Diagn Res. 2013;7:1328-31.
- [Google Scholar]
- Multidrug resistant NDM1 metallobetalactamase producing Klebsiella pneumoniae sepsis outbreak in a neonatal intensive care unit in a tertiary care center at central India. Indian J Pathol Microbiol. 2014;57:65-8.
- [Google Scholar]
- Microbiological evaluation of catheter associated urinary tract infection in a tertiary care hospital. People J Sci Res. 2015;8:23-9.
- [Google Scholar]
- Catheter associated urinary tract infections (CAUTI) and antibiotic sensitivity pattern from confirmed cases of CAUTI in a tertiary care hospital: A prospective study. Clin Microbiol. 2015;4:193.
- [Google Scholar]
- Catheter associated urinary tract infection (CAUTI)incidence and microbiological profile in a tertiary care hospital in Andhra Pradesh. Indian J Microbiol Res. 2016;3:453-6.
- [Google Scholar]
- Prevalence of microorganisms causing catheter associated urinary tract infections (CAUTI) among catheterized patients admitted in a tertiary care hospital. Int J Res Med Sci. 2017;5:2367-72.
- [Google Scholar]
- Molecular characterization and antimicrobial susceptibility profile of New Delhi metallobetalactamase-1-producing Escherichia coli among hospitalized patients. J Lab Physicians. 2018;10:149-54.
- [Google Scholar]
- Incidence of bla NDM1 gene in Escherichia coli isolates at a tertiary care referral hospital in Northeast India. Indian J Med Microbiol. 2013;31:250-6.
- [Google Scholar]
- Statewide surveillance of carbapenemresistant Enterobacteriaceae in Michigan. Infect Control Hosp Epidemiol. 2014;35:3429.
- [Google Scholar]
- Novel variant NDM-11 and other NDM-1 variants in multidrug-resistant Escherichia coli from South India. Journal of global antimicrobial resistance. 2018;14:154-7.
- [Google Scholar]
- Molecular epidemiology of betalactamase producing nosocomial gram-negative pathogens from North and South Indian hospitals. J Med Microbiol. 2017;66:999-1004.
- [Google Scholar]





