Translate this page into:
Antimicrobial Susceptibility Trends among Pathogens Isolated from Blood: A 6-Year Retrospective Study from a Tertiary Care Hospital in East Sikkim, India
Address for correspondence Tsering Yangzom, MBBS, MD (Microbiology), Department of Microbiology, Sikkim Manipal Institute of Medical Sciences, Sikkim Manipal University, 5th mile, Tadong, Sikkim 737102, India (e-mail: tsering.y@smims.smu.edu.in).
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Bloodstream infections (BSIs) are one of the frequent nosocomial infections among hospitalized patients. To understand the local epidemiology and evolving antimicrobial drug resistance of blood-borne pathogens, we analyzed the distribution and antibiotic sensitivity profile of organisms causing BSI in our hospital-based study.
Materials and Methods
We reviewed retrospective data of laboratory-confirmed BSIs, from January 2013 to December 2018. Causative organisms and their antibiotic susceptibility profile of primary and secondary BSI reports were determined from BacT/Alert and Vitek systems findings (bioMérieux). A 6-year multidrug resistance indexing was done to document the resistance pattern of the commonly isolated organisms.
Results
A total of 1,340 (10.2%) BSIs were reported from 13,091 blood cultures. Organisms were frequently isolated from the younger population (≤20 years), especially from ages < 1 year (20.8% of total BSIs). Majority of pathogens were bacterial (97.1%) whereas 2.9% were fungal in origin. Monomicrobial growth was recorded in over 98% of BSIs. Gram-positive and gram-negative bacteria isolated were 518 (39.8%) and 783 (60.2%), respectively. Commonly isolated organisms were coagulase-negative Staphylococci (29.4%), Escherichia coli (19.8%), Klebsiella species (13.5%), Salmonella species (9.4%), and Staphylococcus aureus (7.5%). Multidrug-resistance index was observed highest in Acinetobacter species followed by Pseudomonas aeruginosa and S. aureus.
Conclusion
Overall, there has been a gradual decline in the reporting of BSI. However, infections by gram-negative bacilli and multidrug-resistant organisms remain persistently high. Ages < 20 years were the vulnerable group, with infants < 1 year contributing to the maximum number of BSI cases caused by both bacteria and fungi. Therefore, additional methods are required to study the origin and causation of these infections, particularly among vulnerable patients.
Keywords
antimicrobial drug resistance
drug resistance
bloodstream infection
nosocomial infections
Introduction
Episodes of bloodstream infection (BSI) is a cause of grave concern to a health care facility. BSI complicated by drug-resistant organisms are known to cause increased morbidity, mortality, and hospital expenditure.1-3 The clinical and epidemiological impact of such antimicrobial resistance (AMR) is a worrisome trend posing severe limitations and challenges to clinicians and health care policymakers alike.46
In the epidemiological triad (organism, host, and environment) of BSI, the role of infecting organism is often the most complex to determine, and stringent identification based upon varying parameters is of utmost importance in distinguishing a commensal flora from a pathogen.7 Accordingly, adherence to guidelines helps in avoiding unnecessary treatment of colonization in the absence of infection. Although antibiotic usage exerts selective pressure on the selection of AMR organisms, it is the poor/lack of infection prevention and control practices that facilitate the dissemination of these organisms.4
The AMR among pathogens, particularly in a hospital setting, is increasing and variations in local trends and epidemiology of BSI require constant surveillance.2,8,9 The study aims to address the paucity of information regarding the AMR status of blood-borne pathogens from this region. The objective of this study is to understand the local epidemiology of BSI and the status of antibiotic resistance by evaluating the distribution and AMR of the blood-borne organisms isolated over a 6-year duration.
Materials and Methods
Study Design
This study is a retrospective chart review of blood culture records of patients admitted to a 500-bedded referral hospital in Sikkim, which caters to a semi-urban and rural Himalayan population, from January 2013 to December 2018. This study was reviewed and approved by the Institutional Ethics Committee.
Data Collection and Interpretation
Electronic and manual records of laboratory-confirmed BSI (LCBSI) reports were analyzed and the collected data were systematized by their year of isolation, the demography of patients, and the antibiotic susceptibility profile.
Definition of LCBSI of primary (blood-borne) and secondary origins (another site-specific infection) is based upon the guidelines provided by the Centre for Disease Control and Prevention.7 To avoid repetitive sampling error from a patient having > 1 similar isolate (≤14 days apart) only the first isolate was included. If a patient had ≥ 2 BSI occurring 30 days after the first isolation, each BSI was considered as a new infection.2 Polymicrobial growth is defined as the growth of > 1 clinically relevant organism from single or different blood cultures collected within 48 hours from the first collection.2
Identification and Antibiotic Susceptibility Tests
Processing of blood samples was done in the Department of Microbiology. Aseptically collected paired blood samples were inoculated in BacT/Alert FAN Plus media bottles (adult and pediatric bottles accordingly) and incubated in a BacT/Alert systems (bioMérieux). Positive BacT/Alert blood samples were further cultured in blood agar and MacConkey agar. Final identification and antibiotic susceptibility tests (AST) were done in a Vitek 2 systems (bioMérieux) using Vitek 2 Identification and AST cards for gram-positive or gram-negative organisms, according to the Clinical and Laboratory Standards Institute (CLSI)-defined interpretive minimum inhibitory concentration breakpoints.10,11
Multidrug Resistance Index
Calculated as per the formula described by Krumperman: a/(b × c)12 where, the cumulative antibiotic resistance of all isolates from blood (a) is divided with the denominator, which is the number of antibiotics used (b) multiplied with the number of isolates from blood culture (c).
Multidrug resistance index (MDRI) of < 0.2 and > 0.2 is taken as an indicator to differentiate between low and high-risk source of resistance, respectively.
Organisms such as Acinetobacter baumannii complex, Pseudomonas aeruginosa, Klebsiella species, and Enterococcus species have an innate resistance to certain antibiotics.11 These intrinsically resistant antibiotics were excluded from the index calculation.
Statistical Analysis
The collected data were prepared using MS Excel (Microsoft) spreadsheets. Continuous variables are represented as mean (with standard deviation) and categorical data are expressed in numbers/percentages. Statistical calculations were done using SPSS Statistics version 20.0 (IBM Inc.). Significant p-level < 0.05 was considered.
Results
Over a period of 6 years, 13,091 blood samples were cultured and 1,340 (10.2%) positive BSIs were reported. ►Fig. 1 represents the year-wise distribution of BSI isolates. ►Table 1 shows the distribution of organisms; the majority of the isolates were from medicine wards (58.9%), followed by intensive care unit (ICU) patients under medicine (11.2%) and pediatrics (10.2%). Fungal isolates were from patients admitted under neonatal intensive care unit and pediatric intensive care unit (53.8%). The maximum bacterial BSIs (32.1%, n = 418) were reported from patients < 20 years of age. Patients < 1 year of age constituted 20.8% (n = 279) of total positive samples, whereas, 79.2% (n = 1,061) were of mean age 43.7 ± 21.5 (SD) years. Male to female ratio was 1.09:1.
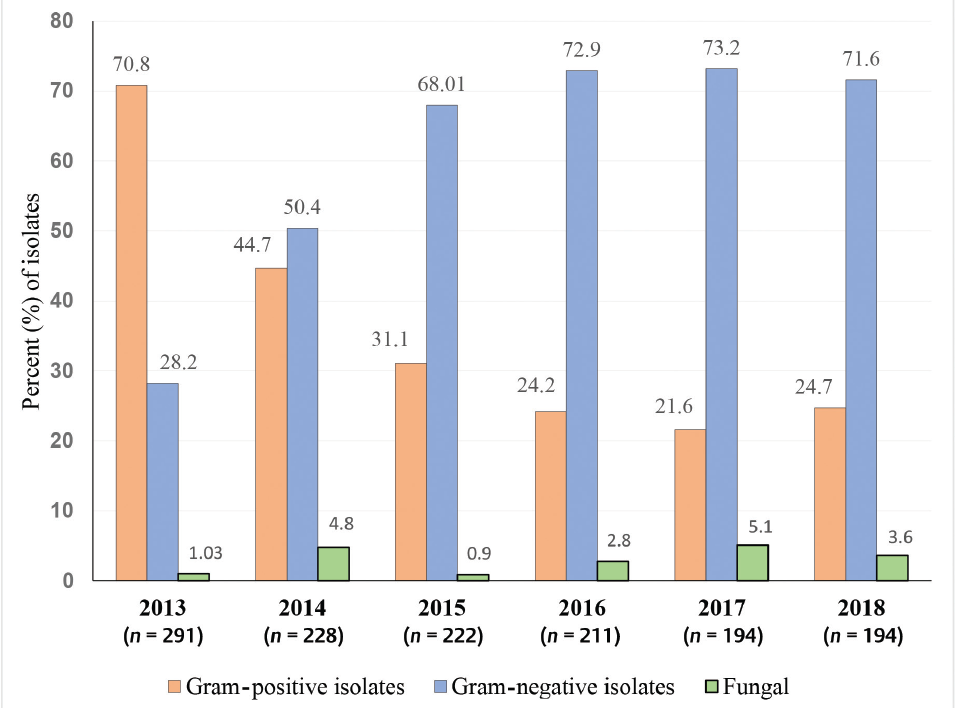
- Percentage distribution of organisms isolated from positive blood culture (n = 1,340) over a duration of 6 years.
| Location | Total bacterial isolates n = 1,301 | Total fungal isolates n = 39 | ||||
|---|---|---|---|---|---|---|
| Gram-positive isolates n = 518 | Gram-negative isolates n = 783 | |||||
| Medicine wards | 305 (58.9) | 397 (50.7) | 6 (15.4) | |||
| Pediatric wards | 44 (8.5) | 22 (2.8) | Not recorded | |||
| Surgical wards | 15 (2.9) | 35 (4.5) | Not recorded | |||
| Private wards | 15 (2.9) | 22 (2.8) | Not recorded | |||
| Obstetrics | 3 (0.6) | 1 (0.1) | Not recorded | |||
| Orthopedics | 1 (0.2) | Not recorded | Not recorded | |||
| NICU and PICU | 53 (10.2) | 204 (26.1) | 21 (53.8) | |||
| MICU and CCU | 58 (11.2) | 55 (7.0) | 10 (25.6) | |||
| Surgical ICU | 11 (2.1) | 34 (4.3) | 2 (5.1) | |||
| Dialysis | 13 (2.5) | 13 (1.7) | Not recorded | |||
| Total bacterial isolates n = 1301 | Gram-positive isolates n = 518 | Gram-negative isolates n = 783 | p-Value | Total fungal isolates n = 39 | ||
| Age in years | IQRa | 45 (58-13) | 39 (58-19) | 57 (58-1) | 48 (49-1) | |
| < 1 to 20 | 418 (32.1) | 144 (27.8) | 274 (34.9) | 0.007a | 21 (53.8) | |
| 21–40 | 335 (25.7) | 139 (26.8) | 196 (25) | 0.47 | 7 (17.9) | |
| 41–60 | 272 (20.9) | 126 (24.3) | 146 (18.6) | 0.01a | 6 (15.4) | |
| > 61 | 276 (21.2) | 109 (21.04) | 167 (21.3) | 0.9 | 5 (12.8) | |
| Sex | Male | 679 (52.2) | 277 (53.5) | 402 (51.3) | 0.45 | 20 (51.3) |
| Female | 622 (47.8) | 241 (46.5) | 381 (48.7) | 19 (48.7) | ||
Abbreviations: CCU, cardiothoracic care unit; ICU, intensive care unit; IQR, interquartile range; MICU, medical intensive care unit; NICU, neonatal intensive care unit; PICU, pediatric intensive care unit.
Note: Figures in parentheses are in percentages.
aIQR, Interquartile range expressed in years; parentheses for upper and lower quartile. Significant value (p ≤0.05).
Secondary BSI in 14.02% of patients had comparable organisms isolated from the respiratory tract (122, 64.8%), urine (36, 19.1%), and pus (30, 15.6%). Significant polymicrobial growth constituted 1.6% (n = 21) of LCBSI organisms.
Most common isolates among the gram-positive bacteria were coagulase-negative Staphylococci (CoNS; 383, 73.9%), followed by Staphylococcus aureus (97, 18.7%), Enterococcus species (30, 5.7%), and Streptococcus species (8, 1.5% [two Streptococcus pyogenes; three Streptococcus agalactiae; one each of Streptococcus pneumoniae, Streptococcus anginosus, and Aerococcus species]).
Most common isolates among the gram-negative bacteria were Escherichia coli (257, 32.8%), followed by Klebsiella species (175, 22.3%), Salmonella species (122, 15.2%), Acinetobacter species (54, 6.9%), Pseudomonas species (48, 6.1%), Burkholderia cepacia (32, 4.1%), Enterobacter species (26, 3.3%), and Serratia species (15, 1.9%). Other less frequently isolated organisms were Sphingomonas species (8, 1.02%), Proteus species (8, 1.02%), Citrobacter species (7, 0.9%), Aeromonas species (5, 0.6%), Achromobacter species (4, 0.5%), Morganella species (4, 0.5%), Cronobacter sakazakii (3, 0.4%), and one (0.1%) isolate each of Elizabethkingia species, Chryseobacterium species, Shigella species, and Alcaligenes faecalis.
The most common organisms isolated from the ICUs and the wards were Klebsiella species (n = 122) and CoNS (n = 298), respectively. On evaluation of organism’s likelihood of isolation from patients in the ICUs or the wards (odds ratio [OR] 95% confidence interval [CI]), Klebsiella species (OR: 6.5, CI: 4.6–9.2, p < 0.0001), Enterobacter species (OR: 6.0, CI: 2.5–14.3, p < 0.0001), Enterococcus species (OR: 2.5, CI: 1.2–5.1], p = 0.02), Acinetobacter species (OR: 1.9, CI: 1.1–3.3, p = 0.03), and Pseudomonas species (OR: 2.0, CI: 1.1–3.6, p = 0.02) were likely from the ICU patients whereas CoNS, S. aureus, E. coli, and Salmonella species were more likely from the ward patients.
►Table 2 represents the antibiotic resistance of the commonly isolated gram-negative bacteria. E. coli, Klebsiella, and Enterobacter species had similar resistance pattern with > 60% resistance to ampicillin, cefuroxime, ceftriaxone, and cefepime. Acinetobacter species displayed high resistance to almost all antibiotics tested, except, tigecycline and colistin. Salmonella paratyphi A was the frequent isolate among Salmonella species (59%). Only nalidixic acid-resistance (84.4%) was prominent among the Salmonella species.
| Antimicrobials | E. coli n = 257 | Klebsiella species n = 175 | Salmonella species n = 122 | Acinetobacter species n = 54 | Pseudomonas species n = 48 | Enterobacter species n = 26 |
|---|---|---|---|---|---|---|
| AMP | 196 (76.3) | 167 (95.4) | 12 (9.8) | 49 (90.7) | 42 (87.7) | 19 (73.1) |
| AMC | 95 (36.9) | 78 (44.6) | 4 (3.3) | 46 (85.2) | 33 (68.6) | 23 (88.5) |
| PTZ | 42 (16.3) | 43 (24.6) | 4 (3.3) | 32 (59.3) | 17 (35.4) | 4 (15.4) |
| 2nd gen Ceph | 192 (74.7) | 148 (84.6) | NRa | 48 (88.9) | 42 (87.7) | 24 (92.3) |
| CTX | 171 (66.5) | 138 (78.9) | 10 (8.2) | 43 (79.6) | 34 (70.8) | 18 (69.2) |
| CF-SUL | 32 (12.5) | 37 (21.1) | 2 (1.6) | 23 (42.6) | 20 (41.7) | 5 (19.2) |
| CPM | 157 (61.1) | 141 (80.6) | 9 (7.4) | 39 (72.2) | 24 (50) | 10 (38.5) |
| AMK | 14 (5.4) | 30 (17.1) | NRa | 30 (55.6) | 16 (33.3) | 3 (11.5) |
| GEN | 81 (31.5) | 91 (52) | NRa | 31 (57.4) | 21 (43.8) | 15 (57.7) |
| NA | 180 (70) | 70 (40) | 103 (84.4) | 32 (59.3) | 34 (70.8) | 8 (30.8) |
| CIP | 148 (57.6) | 52 (29.7) | 33 (27) | 28 (51.9) | 26 (54.2) | 4 (15.4) |
| TIG | 23 (8.9) | 22 (12.6) | 2 (1.6) | 11 (20.4) | 30 (62.5) | 3 (11.5) |
| Carbapenem | 12 (4.7) | 17 (9.7) | 2 (1.6) | 22 (40.7) | 14 (29.2) | 4 (15.4) |
| CoT | 146 (56.8) | 96 (54.8) | 10 (8.2) | 34 (62.9) | 37 (77.1) | 9 (34.6) |
| COL | 16 (6.2) | 9 (5.1) | 5 (4.1) | 10 (18.5) | 4 (8.3) | 3 (11.5) |
Abbreviations: 2nd gen Ceph, 2nd gen cephalosporin; AMC, amoxycillin-clavulanic; AMK, amikacin; AMP, ampicillin; CF-SUL, cefoperazone/sulbactam; CIP, ciprofloxacin; COL, colistin; CoT, trimethoprim-sulfamethoxazole; CPM, cefepime; CTX, ceftriaxone; GEN, gentamicin; NA, nalidixic acid; NR, not reported; PTZ, piperacillin/tazobactam; TIG, tigecycline.
Note: Values in parentheses are in percentages. Second generation cephalosporin includes cefuroxime and cefuroxime axetil; Carbapenem includes ertapenem, imipenem, and meropenem.
aAccording to CLSI, Salmonella spp. susceptibility to second generation cephalosporin and aminoglycoside should not be reported as they may appear active in vitro but are ineffective clinically.11
Antibiogram of gram-positive organism indicates high resistance to benzylpenicillin, erythromycin, and ciprofloxacin as represented in ►Table 3. Vancomycin-resistance was lower in Staphylococcus species than Enterococcus species. Among enterococci, vancomycin resistance was seen only in Enterococcus faecalis (80%) and Enterococcus faecium (20%), whereas high-level gentamicin (HLG) resistance was observed higher in E. faecium (53.8%) than in E. faecalis (46.1%). Enterococcus casseliflavus and Enterococcus gallinarum isolates were vancomycin and HLG-sensitive isolates.
| Antimicrobials | Coagulase negative Staphylococci n = 383 | Staphylococcus aureus n = 97 | Enterococcus species n = 30 |
|---|---|---|---|
| PEN | 326 (85.1) | 81 (83.5) | 19 (63.3) |
| OXA | 222 (57.9) | 43 (44.3) | Not done |
| CIP | 126 (32.9) | 40 (41.2) | 20 (66.7) |
| LFLX | 99 (25.8) | 37 (38.1) | 16 (53.3) |
| GEN | 46 (12.0) | 12 (12.4) | Not done |
| HLG | Not done | Not done | 13 (43.3) |
| ERY | 230 (60.1) | 44 (45.4) | 20 (66.7) |
| CD | 130 (33.9) | 27 (27.8) | Not reporteda |
| TET | 39 (10.2) | 5 (5.2) | 11 (36.7) |
| CoT | 135 (35.2) | 43 (44.3) | Not reporteda |
| VAN | 7 (1.8) | 6 (6.2) | 5 (16.7) |
| TEI | 14 (3.7) | 5 (5.2) | 4 (13.3) |
| LIN | 9 (2.3) | 0 | 0 |
| DAP | 4 (1.0) | 2 (2.1) | 0 |
Abbreviations: CD, clindamycin; CIP, ciprofloxacin; CoT, trimethoprim-sulfamethoxazole; DAP, daptomycin.; ERY, erythromycin; GEN, gentamicin; HLG, high-level gentamicin; LFLX, levofloxacin; LIN, linezolid; OXA, oxacillin; PEN, benzylpenicillin; TEI, teicoplanin; TET, tetracycline; VAN, vancomycin.
Note: Figures in parentheses are in percentages.
aAccording to CLSI, Enterococcus spp. susceptibility to low-level aminoglycoside, clindamycin, and co-trimoxazole should not be reported as they may appear active in vitro but are ineffective clinically.11
Fungi constituted 2.9% of positive cultures. They were caused by yeast-like organisms such as Candida albicans and Candida tropicalis. Candida species were commonly isolated from ICU patients (82.1%) with a mean age of 41.9 ± 23.7 (SD) years and a male to female ratio of 1.05:1. Neonates contributed to 43.6% of total fungal BSI (►Table 1).
On the year-wise evaluation of MDRI of the commonly isolated organism for a duration of 6 years, it is evident that there is a significant rise in resistance pattern of all isolates, particularly A. baumannii complex, P. aeruginosa, and S. aureus (►Fig. 2). A. baumannii complex and P. aeruginosa had the highest number of multidrug resistances throughout the duration of the study period.
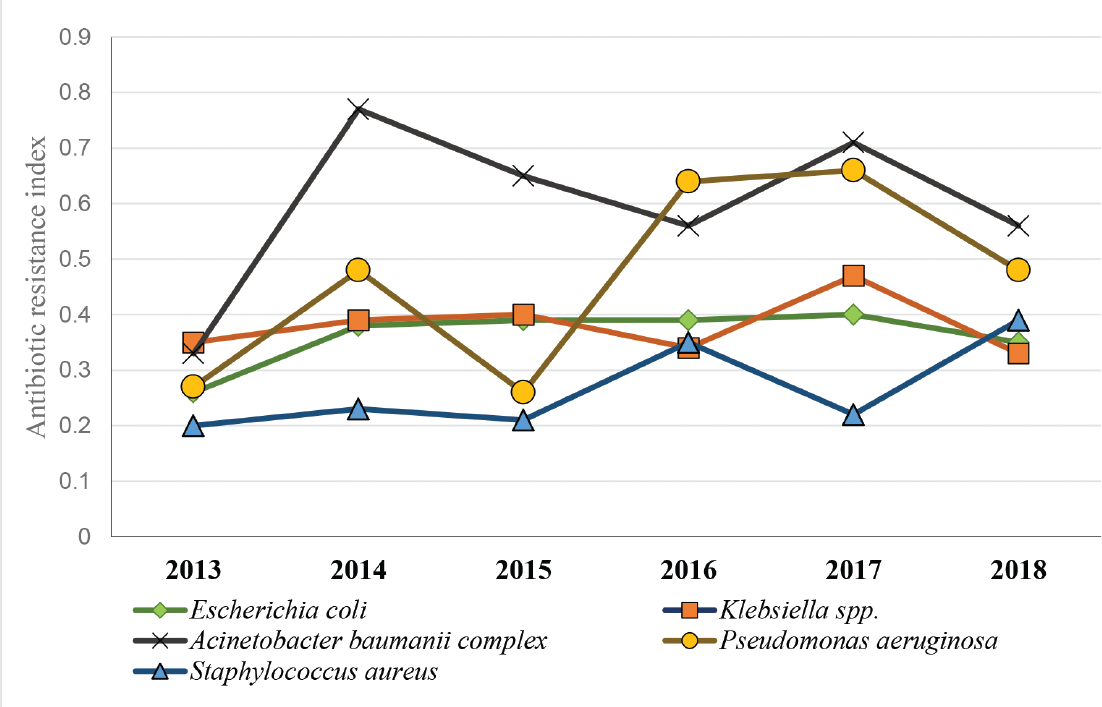
- Multiple antibiotic resistance index of commonly isolated organisms from bloodstream infections over a duration of 6 years.
Discussion
Similar to several studies, gram-negative bacilli were the common isolates (60.2%) in our study.8,13-15 Gram-positive cocci were the commonly isolated organisms at the beginning of our study in 2013, with CoNS as the frequently isolated group.16 However, by 2014 onward, upward trend of gram-negative bacilli isolation increased when compared with a decline in the frequency of CoNS isolation. Overall, a gradual decrease in isolation of both types of organisms, with gram-positive cocci more so, could perhaps be attributed to a better understanding of sample collection, infection control, and prevention.9,13
In our study, the rise in total antibiotic resistance of S. aureus is of concern. Till 2017, S. aureus had the lowest MDR index values, but by 2018, it had abruptly risen to become the organism with the third-highest values. This rise could perhaps be attributed to the majority of isolates developing resistance to many of the first-line antibiotics such as fluoroquinolones, erythromycin, and penicillin.9,14 On the contrary, few gram-positive cocci in our study were vancomycin-resistant, which is similarly reported by Zhu et al.9
Similar to the findings by Djuric et al, the overall resistance among gram-negative isolates was observed highest to β-lactams, trimethoprim-sulfamethoxazole, and aminoglycosides.13 The MDRI of commonly isolated gram-negative bacilli was alarmingly high and remained consistently high throughout our study period. The resistance pattern is comparable to reports indicating a predictable rise of MDR organisms in the future.4,6
Carbapenem resistance in our study was highest for Acinetobacter species (40.7%), also reported by Kumar et al.15 Carbapenem resistance of > 20% was detected in Pseudomonas species and Burkholderia species, whereas the lowest resistance was among the Enterobacteriaceae, which is in contrast to the study by Datta et al17 Nevertheless, close monitoring and improvement of infection control should be implemented to avoid the emergence of carbapenem-resistant Enterobacteriaceae (CRE).4 All the gram-negative isolates in our study were mostly sensitive to colistin, which is the drug of choice in treating CRE. However, according to the recent CLSI guidelines, broth microdilution is the only acceptable method for detecting colistin resistance and Vitek 2 GN-AST cards use agar dilution method for antibiotic sensitivity testing.11 Consequently, the importance of the colistin susceptibility reports from our study remains uncertain.
Though the MDR indices of E. coli and Klebsiella species were fairly constant, it was consistently high throughout the duration of the study. E. coli was the most common gram-negative organism causing BSI in our study and it is also implicated as a frequent organism causing BSI in Europe and in the SENTRY surveillance program.4,18
In our study, the MDRI of A. baumannii complex has been the highest. It exhibited resistance to mostly all the antibiotics, except tigecycline and colistin, which is also reported elsewhere.13,15 Surveillance studies in Europe have revealed that Acinetobacter species have an alarmingly high level of AMR to routinely used antibiotic groups of fluoroquinolones, aminoglycosides, and carbapenems.4
Though the isolation rate of Pseudomonas species in this study is less (3.6%), its resistance to many antibiotics (excluding colistin) increased significantly over 6 years. P. aeruginosa is an organism responsible for the majority of health care-associated infections, with its intrinsic and acquired resistance to several antibiotics adding pressure to a preexisting global AMR burden.4
The current situation of BSI in India is rather complex to quantify as regional variations abound.14 The Centers for Disease Control and Prevention and SCOPE project (Surveillance and Control of Pathogens of Epidemiological Importance) in the United States monitor BSI via a nationwide network of hospitals, similar to the SENTRY Antimicrobial Surveillance Program, the European Antimicrobial Resistance Surveillance Network, Central Asian and Eastern European Surveillance of Antimicrobial Resistance network, and the Asian-Pacific Research Foundation for Infectious Diseases of South Korea.4,7,13,18,19 The SCOPE nationwide collective effort is also adopted by Brazil.8 Such surveillance programs are the need of the hour in India as there are inadequate databases and no repository of resistant pathogens that streamline active surveillance programs (to guide clinical decisions on the use of antimicrobials).20 Hopefully, the Antimicrobial Resistance Surveillance and Research Network and National Action Plan on Antimicrobial Resistance will provide some solutions in tackling the problem of emerging AMR pathogens collective for the Indian subcontinent.20
Limitations
The study has several limitations. First, the data that were dependent upon the medical records were inconsistent with the timings of hospital admissions, due to which BSI could not be classified as either hospital or community acquired. Second, due to the retrospective nature of this study, many isolates could not be assessed for molecular studies to evaluate the genetic determinants of AMR. Finally, BSI caused by anaerobic organisms and fungal antibiotic sensitivity testing is not done routinely in our hospital, therefore, the prevalence of anaerobic BSI and fungal antibiogram is unknown.
Conclusion
A progressive increase in multidrug (≥3 antibiotics)-resistant organisms is posing several challenges to our hospital infection-control operations. More studies are required to comprehend the etiology of such AMR organisms to aid appropriate antibiotic usage, control, and contain the spread of AMR organisms in the future.
Institutional Research Protocol Evaluation Committee Review
Approved by IRPEC on June 18, 2019 under SMIMS IRPEC Registration No: IRPEC/398/19–067.
Contribution of Authors
T.Y. is responsible for the study conception, design, collection of data, drafting, and final revision of the manuscript. D.C.T. is responsible for the study conception, drafting, and final revision of the manuscript. S.K. is responsible for result analysis, interpretation, and critical revision of the manuscript. J.K. is responsible for the drafting and critical revision of the manuscript.
Conflict of Interest
The authors declare that there is no conflict of interest.
Funding
None
Reference
- Nosocomial bloodstream infections in United States hospitals: a three-year analysis. In: Clin Infect Dis. Vol 29. 1999. p. :239-244.
- [CrossRef] [PubMed] [Google Scholar]
- Ten-year surveillance of nosocomial bloodstream infections: trends of aetiology and antimicrobial resistance in a comprehensive cancer centre. Ecancermedicalscience. 2011;5:191.
- [Google Scholar]
- Educational interventions for preventing vascular catheter bloodstream infections in critical care: evidence map, systematic review and economic evaluation. Health Technol Assess. 2014;18(15):1-365.
- [CrossRef] [Google Scholar]
- Surveillance of antimicrobial resistance in Europe—Annual Report of the European Antimicrobial Resistance Surveillance Network. 2019 Available at: https://ecdc.europa.eu/sites/portal/files/documents/EARS-Net-report-2017-update-jan-2019.pdf. Accessed February 2, 2019
- [Google Scholar]
- The increasing challenge of multidrug-resistant gram-negative bacilli. Results of a 5-year active surveillance program in a neonatal intensive care unit. Medicine (Baltimore). 2016;95(10):e3016.
- [CrossRef] [PubMed] [Google Scholar]
- Ministry of Health and Family Welfare, Government of India. 2017 Available at: http://www.searo.who.int/india/topics/antimicrobial_resistance/nap_amr.pdf. Accessed March 26, 2019
- [Google Scholar]
- Bloodstream Infection Event (Central Line-Associated Bloodstream Infection and Non-central Line Associated Bloodstream Infection) Atlanta, GA: CDC; 2019. Available at: https://www.cdc.gov/nhsn/PDFs/pscManual/4PSC_CLABScurrent.pdf. Accessed April 14, 2019
- [Google Scholar]
- Brazilian SCOPE Study Group. Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J Clin Microbiol. 2011;49(05):1866-1871.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology and microbiology of gram-positive bloodstream infections in a tertiary-care hospital in Beijing, China: a 6-year retrospective study. Antimicrob Resist Infect Control. 2018;7:107.
- [CrossRef] [PubMed] [Google Scholar]
- CLSI supplement M100S. (26th). Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
- [Google Scholar]
- CLSI supplement M100. (29th). Wayne, PA: Clinical and Laboratory Standards Institute; 2019.
- [Google Scholar]
- Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(01):165-170.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial resistance of selected invasive bacteria in a tertiary care center: results of a prospective surveillance study. J Infect Dev Ctries. 2016;10(12):1325-1331.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence, etiology, and antibiotic resistance profiles of bacterial bloodstream infections in a tertiary care hospital in Northern India: a 4-year study. J Lab Physicians. 2018;10(04):426-431.
- [CrossRef] [PubMed] [Google Scholar]
- Risk factors for carbapenem-resistant Acinetobacter baumanii blood stream infections in a neonatal intensive care unit, Delhi, India. J Infect Dev Ctries. 2014;8(08):1049-1054.
- [CrossRef] [PubMed] [Google Scholar]
- Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008-2014. Int J Infect Dis. 2016;50:75-82.
- [CrossRef] [PubMed] [Google Scholar]
- A ten year analysis of multi-drug resistant blood stream infections caused byEscherichia coli&Klebsiella pneumoniaein a tertiary care hospital. Indian J Med Res. 2012;135(06):907-912.
- [Google Scholar]
- The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. 2019;63(07):e00355-19.
- [CrossRef] [PubMed] [Google Scholar]
- Bloodstream infections and clinical significance of healthcare-associated bacteremia: a multicenter surveillance study in Korean hospitals. J Korean Med Sci. 2010;25(07):992-998.
- [CrossRef] [PubMed] [Google Scholar]
- Fostering research into antimicrobial resistance in India. BMJ. 2017;358:j3535.
- [CrossRef] [PubMed] [Google Scholar]






