Translate this page into:
Clinical spectrum and diagnostic yields of Mycoplasma pneumoniae as a causative agent of community-acquired pneumonia
Address for correspondence: Dr. Rama Chaudhry, Department of Microbiology, AIIMS, New Delhi, India. E-mail: drramach@gmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
INTRODUCTION:
Infection with Mycoplasma pneumoniae (M. pneumonia) occurs worldwide which accounts for 15%–20% of cases of community-acquired pneumonia and indistinguishable clinically from other infectious causes of pneumonia.
AIM:
The aim of this study was to evaluate the real-time polymerase chain reaction (PCR) and to correlate it with other diagnostic methods such as culture, serology (ELISA), and conventional PCR along with the clinical signs and symptoms produced by M. pneumonia.
MATERIALS AND METHODS:
A total of 130 patients of all age groups presenting with clinical features of lower respiratory tract infections were enrolled over a period of 1 year and 2 months in a tertiary care hospital in Delhi. M. pneumoniae in throat swab samples was detected by real-time PCR, compared with culture, serology, conventional PCR, and clinical signs and symptoms. Univariate analyses were conducted to determine the association of M. pneumoniae infection among different categories of patients.
RESULTS:
Out of a total of 130 patients, 18 patients (14%) were positive for M. pneumoniae by any test; culture was positive in nine patients (50%), serology (IgM) in eight patients (44.4%), PCR in five patients (27.7%), and real-time PCR was positive in six patients (33.3%). Clinical signs and symptoms were higher in incidence in M. pneumoniae-positive patients. Age-matched healthy controls (30) were included in the study, and all were negative for any diagnostic test performed (P = 0.026).
CONCLUSION:
It was concluded that combination of M. pneumoniae-specific testing modalities is required for the diagnosis of this etiological agent rather than a single diagnostic method.
Keywords
Community-acquired pneumonia - Mycoplasma pneumoniae
polymerase chain reaction
real-time polymerase chain reaction
serology
Introduction
Mycoplasma pneumoniae (M. pneumonia) is an important etiological agent of community-acquired pneumonia (CAP) in children as well as in the elderly age groups; the prevalence being highest among school-aged children of 5–15 years of age.[12] M. pneumoniae is principally transmitted by droplet infection from person to person or by fomites to close contacts. It infects both the upper and lower respiratory tracts, leading to upper respiratory tract infection, tracheobronchitis, bronchitis, bronchiolitis, and CAP.[3] The most common manifestations include sore throat, fever, cough, headache, chills, coryza, myalgia, earache, and general malaise.[4]
Pneumonia caused by this agent mimics viral pneumonia[5] which may account for 15%–20% of cases of CAP with up to 18% hospitalization rates in children.[6] Information regarding pneumonia caused by M. pneumoniae is scarce in Indian setup which may be due to under reporting or due to lack of a suitable diagnostic method. A number of diagnostic methods such as culture, complement fixation test, and serology is available, but each of these methods has their own limitations.[78] Recently, molecular diagnosis by real-time polymerase chain reaction (PCR) has gained its popularity due to its high sensitivity and specificity. Hence, the present study was conducted to evaluate the real-time PCR and to correlate with other diagnostic methods such as culture, serology (ELISA), and conventional PCR along with the clinical signs and symptoms of pneumonia caused by M. pneumoniae.
Materials and Methods
Study design
After obtaining the Ethical Committee approval (ethical clearance letter no - International Ethics Standards Coalition/T-13/30.12.11), this prospective study was conducted over a period of 1 year and 2 months (from February 2012 to April 2013) among patients of all age groups admitted to the medicine and pediatric department (both outpatients and inpatients) of All India Institute of Medical Sciences, New Delhi. Healthy volunteers without any apparent disease were taken as control group. Blood and throat swab samples were collected from both the groups of patients. Blood was taken for serology and throat swab for culture, PCR, and real-time PCR.
Inclusion criteria
All patients (both pediatrics and elderly age group) having definitive radiological evidence of pneumonia were included along with at least two of the following clinical features (1) fever, (2) cough, (3) coryza/sore throat, (4) wheeze, and (5) breathlessness.
Exclusion criteria
Patients with terminal illness, hospital-acquired pneumonia, and patients on ventilator were excluded from the study.
Sample
Throat swab and blood samples were collected from 130 patients and 30 controls after their written consent. Throat swab was collected using cotton swabs in sterile containers containing Pleura pneumonia like organism (PPLO) broth with precaution taken not to contaminate the desired sample with oral commensals during collection. Samples were transported in PPLO broth to the laboratory as soon as possible and stored at 4°C. For culture, the samples were inspected daily for color change in PPLO broth for 4 weeks, and change of color to yellow was considered as true-positive culture which was then subcultured on PPLO agar. Serology was performed using Capture ELISA (Institute Virion/Serion, Germany) for detection of IgM and IgG antibodies. For PCR and real-time PCR, genomic DNA was extracted using boiling method. Blood was taken for serology and throat swab for culture, PCR, and real-time PCR. Real-time PCR was performed using TaqMan probe.[9]
Methods
In the previous studies by different authors, PCR has been compared to serology,[1011] and nucleic acid sequence-based amplification has been compared to conventional PCR[12] for the diagnosis of M. pneumoniae infection, but only a few studies have compared the sensitivity of real-time PCR and conventional PCR in patient population. Therefore, we have performed both real-time PCR and PCR to compare their diagnostic yields.
Polymerase chain reaction
For M. pneumoniae P1 adhesin gene, specific primers (published primers) were used.[13] The target sequence for amplification was a 543 bp segment of the gene coding for P1 adhesin protein. The 25 μl PCR reaction consisted of 1X PCR buffer (Bangalore Genei, India), 1.5 mM MgCl2 (Bangalore Genei, India), 200 μM dNTPs (MBI Fermentas, USA), 20 pM of each primer (Sigma-Aldrich, USA), 1 Unit of Taq polymerase (Bangalore Genei, India), and 5 μl of genomic DNA extracted by boiling method.[14]
Forward Primer: 5’CAAGCCAAACACGAGCTCC GGCC3’.
Reverse Primer: 5’GGGGAAGGACAAACAGCTGA CACTGG 3’.
The reaction was performed in a thermocycler (Perkin Elmer, USA). PCR run consisted of 35 cycles of amplification, each at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min and final elongation step of 10 min at 72°C. A negative control was systematically run in parallel. This method was used for PCR amplification from patient samples followed by gel electrophoresis of PCR products [Figure 1].
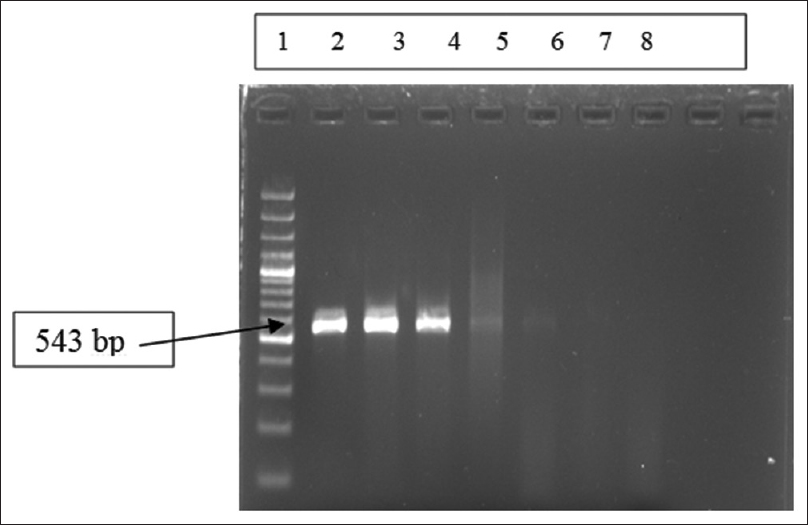
- Gel electrophoresis of polymerase chain reaction amplification products for Mycoplasma pneumoniae P1 gene of 543 bp. Lane 1: Ladder (100 bp ladder); Lane 2–4: Positive controls at different concentration of nucleic acids (copies/μl) used in standardization of real-time polymerase chain reaction, Lane 5–6: Positive samples, Lane-7: Negative sample, Lane 8: Negative control
Real-time polymerase chain reaction standardization
The positive control template was the 543 bp fragment of P1 protein gene cloned in pGEMTE Easy vector. The clone was confirmed by sequencing and used to extract DNA in larger quantity. For this, a 100 mL Luria-Bertani Broth containing ampicillin (100 μg/mL) was inoculated with the glycerol stock. The culture was incubated at 37°C in a shaker (225 rpm/min). The overnight grown culture was harvested, and the plasmid extraction was done using the Qiagen Midi kit. The plasmid DNA extracted was quantified spectrophotometrically.
The copy number calculation of nucleic acid was done using the following formula:
Copy number = 6.023 × 1023 (copies/mol) × Concentration of standard (g/μl)/Molecular weight (g/mol)
Concentration of standard = Obtained spectrophotometrically.
MW = Molecular weight of each pGEM-T Easy plasmid with cloned gene.
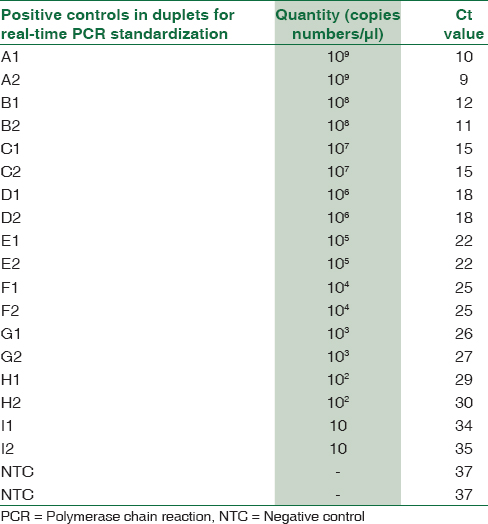
Using above formula, M. pneumoniae P1 gene cloned in pGEM-T Easy vector was calculated to be 1 × 109 copies/μl. A set of dilutions were prepared for standard curve preparation [Table 1]. The primers for real-time assay corresponded to a 73 bp fragment internal to the 543 bp fragment of P1 gene which was detected by Fam-dye labeled probe sequence. Forward Primer 5’ AACCTCGCGCCTAATACTAATACG 3’, Reverse Primer 5’ TTGCGGCGTTGCTTTCAG 3, and Probe Sequence 5’ AAAGTCGACCAACCCC 3’ with FAM label at the 5’ end. The reactions were performed in a final volume of 20 μl containing 1 μl × 20 assay mix (primers and probe), 10 μl of TaqMan Universal Master Mix (enzyme, buffer and dNTPs), and 9 μl DNA diluted in RNase free water. Amplification and product detection were performed with the ABI biosystem 7500 Detection System, USA, and the reaction was carried out in the instrument for 1 h 50 min for the complete amplification. Once the reaction for standard curve was finalized, similar reactions were performed for DNA extracted by boiling method from throat swabs of patients.[14] Cutoff Ct value was taken as 36 and samples showing Ct value above this were considered negative [Table 2].

Results
A total of 130 patients were enrolled in this study, out of which 35 (27%) were females and 95 (73%) were males [Figure 2]. Age wise, number of pediatric and adults patients enrolled were 30 (23%) and 100 (76%), respectively. Healthy volunteers without any apparent disease (n = 30) were taken as control group after written consent. Among control groups, 12 (40%) were females and 18 (60%) were male. Age wise, the number of pediatrics and adults in the control group were 5 (15%) and 25 (85%), respectively [Figure 2]. Enrollment of pediatric population as control group was undertaken after the written consent of their parents.
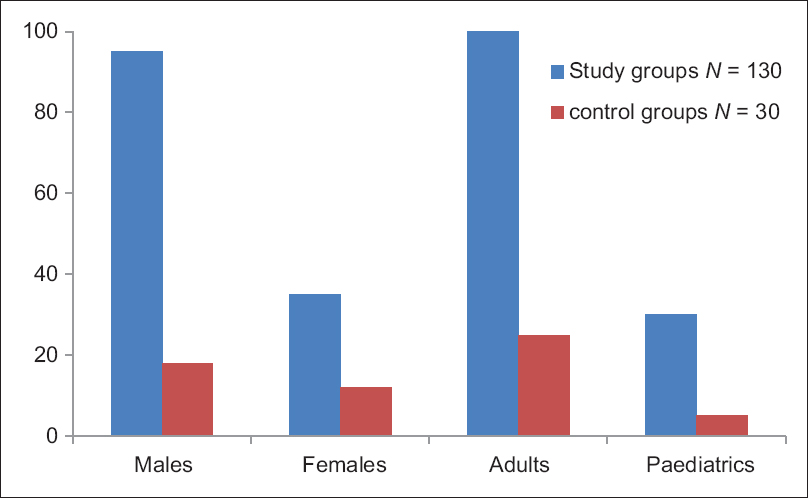
- Showing age and sex distribution of study group and control group
Out of 130 patients, 18 patients (14%) were positive for M. pneumoniae by any test (culture, serology, PCR, and real-time PCR) [Table 3]. Among 18 patients, culture was positive in 9 patients (50%), serology (IgM) in eight patients (44.4%), PCR in five patients (27.7%), and real-time PCR was positive in six patients (33.3%). None of the control groups were positive for any test except IgG antibody which was considered as past infections/recurrent infections. When combinations of different diagnostic modalities were considered [Figure 3], two patients were found to be positive for culture, PCR, and real-time PCR; four patients were positive for both culture and real-time PCR (Negative by conventional PCR); and one patient was positive for culture, serology (IgM), and PCR (negative by real-time PCR) in combination. When age and sex groups were considered, 22% of females, 78% of males, 24% of pediatric patients, and 11% of adults were positive for M. pneumoniae by any test.
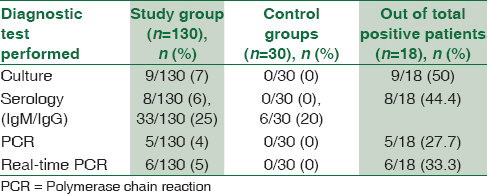
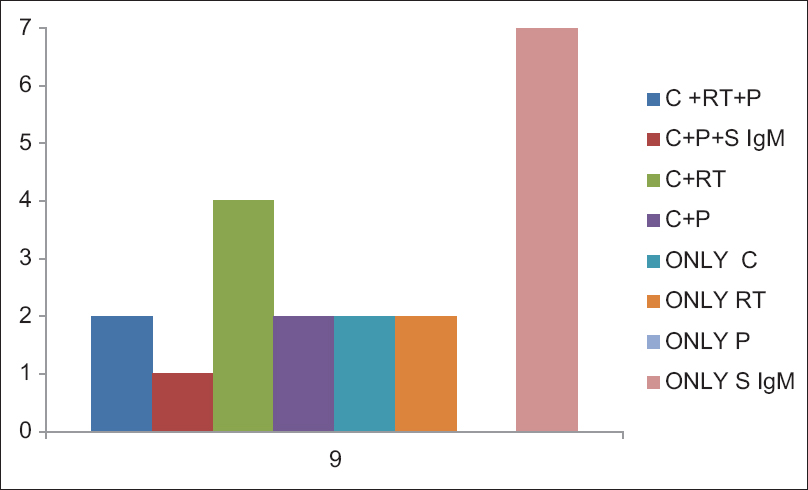
- Figure showing different diagnostic methods in combination which could diagnosed the Mycoplasma pneumoniae infection. Two samples were found to be positive for culture, polymerase chain reaction, and real-time polymerase chain reaction in combination; Four samples were positive for both culture and real-time polymerase chain reaction and one sample was positive for culture, serology (IgM), and polymerase chain reaction in combination; Two patients were positive by only culture, two patients were positive by only real-time polymerase chain reaction, and seven patients were positive by only serum IgM (C = Culture, RT = Real time PCR, P = PCR, S IgM = Serum IgM)
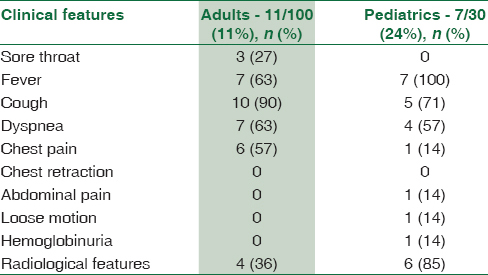
A variety of clinical signs and symptoms were observed in patients suffering from pneumonia [Table 4]. The comparison of clinical data of pneumonia patients with culture, serology, PCR, and real-time PCR [Table 1] revealed that cough and dyspnea were seen in higher proportion in culture-positive cases, whereas fever was present in higher proportion (100%) in real-time PCR and IgM-positive cases. When radiological features were considered, a total of 56 patients (42%) had radiological evidence of pneumonia. Radiological features were higher in real-time PCR-positive cases (84%) which was found to be statistically significant (P< 0.026), followed by PCR-positive cases (80%) and present in near about equal proportion in both culture- and serology-positive samples [Table 1].
Discussion
Most infections with M. pneumoniae are not diagnosed, as they are indistinguishable from upper and lower respiratory tract infections caused by other viral and bacterial pathogens.[15] Many researchers have stated that “No available diagnostic test reliably and rapidly detects M. pneumoniae. Thus, therapy usually is empirical” in the practice guidelines for the management of CAP.[16] M. pneumoniae is difficult to culture also time consuming and needs expertise. Previously, diagnosis usually relied on serology.[17] However, serology is not reliable in specificity, needs paired sera, and is usually positive at about 7 days after the onset of disease.[18] In contrast, molecular tests such as real-time PCR on throat swab and other respiratory secretions may provide an early diagnosis of M. pneumoniae infection and could be used as an useful diagnostic technology.[14]
In the present study, maximum samples were positive by culture, followed by serology and molecular methods. Out of 41 serology-positive samples (IgG + IgM), only IgM-positive samples (n = 8) were considered as true positive [Table 3] because IgG antibody may occur in patients with past infections or recurrent asymptomatic infections. The sensitivity of PCR (27.7%) and real-time PCR (33.3%) was found to be less as compared to other studies; however, still, it is in accordance with other studies,[1920] for example, Otomos et al. have mentioned 6% positivity by real-time PCR. The low sensitivity in our study may be due to the following facts (a) the sensitivity of PCR testing depends on the type of sample tested. Räty et al. reported a sensitivity of 69% in sputum samples, 50% in nasopharyngeal aspirates, and 37.5% in throat swab.[21] (b) P1 cytoadhesin gene has less sensitivity as compared to other targets such as 16 s ribosomal RNA.[22] (c) We used boiling method for extraction of M. pneumoniae DNA, whereas others have used QiaAmp DNA kit.[23] The overall sensitivity of real-time PCR and conventional PCR was very close to each other, but they picked up different samples to be positive, i.e., some real-time PCR-positive samples were not positive by conventional PCR and vice versa.
Most of the patients in our study belonged to indoor patients having severe signs and symptoms. They were already administered two-to-three doses of antibiotics before collection of samples due to some sorts of clinical manifestations which may explain culture negative in serology-positive patients. Out of nine culture-positive patients, eight patients did not show the presence of IgM antibody response which may be due to the fact that the antibody response in M. pneumoniae infection may take 1–2 weeks for development and we collected the specimen at an earlier time leading to no antibody response.
Comparison of clinical signs and symptoms between M. pneumoniae-infected and noninfected patients revealed that cough, dyspnea, chest pain, and radiological features were predominant findings in M. pneumoniae-positive patients [Tables 1 and 4]. Statistically significant difference was obtained for cough and chest pain (P = 0.01 each). When pediatrics and adult age groups were compared, association of sore throat (27%) and chest pain (57%) was found to be higher with the adults, whereas association of radiological features (85%) was found to be statistically significant in the pediatrics age group as compared to adult age group [Table 4].
Conclusion
The present study indicates that there is absence of a single test which can reliably detect M. pneumoniae infection. A combination of two or three methods can be the most reliable approach for identification of CAP due to M. pneumoniae, especially in the absence of other suspected respiratory pathogens. Although the molecular methods reflected a decreased sensitivity for its diagnosis, still, they cannot be ignored specially in the early stage of infections. Boiling method for extraction of DNA may not be a suitable method for molecular diagnosis of M. pneumoniae. Further study is required for the evaluation of a single better diagnostic method.
Financial support and sponsorship
I Thanks ICMR for its financial support.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The funds for this research work were sanctioned by ICMR. The authors sincerely thank to ICMR.
References
- Epidemiology of Mycoplasma pneumoniae infection in families. JAMA. 1966;197:859-66.
- [Google Scholar]
- Introduction to mycoplasma and ureaplasma. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Vol 24. (7th). Philadelphia, PA: Elsevier Churchill Livingstone; 2010. p. :77-81.
- [Google Scholar]
- Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697-728.
- [Google Scholar]
- Respiratory tract infections by Mycoplasma pneumoniae in children: A review of diagnostic and therapeutic measures. Eur J Pediatr. 2001;160:483-91.
- [Google Scholar]
- BTS Guidelines for the Management of Community Acquired Pneumonia in Childhood 2000. Thorax. 2002;57:1-24.
- [Google Scholar]
- New concepts of Mycoplasma pneumoniae infections in children. Pediatr Pulmonol. 2003;36:267-78.
- [Google Scholar]
- Mycoplasma pneumoniae Neurological Disease Associated with a Community Respiratory Outbreak - Rhode Island. Program and abstracts of the 45th Annual Meeting of the Infectious Diseases Society of America. Alexandria, VA: Infectious Diseases Society of America; 2007.
- [Google Scholar]
- Laboratory diagnosis of Mycoplasma pneumoniae infection. Clin Microbiol Infect. 2003;9:263-73.
- [Google Scholar]
- Development of a genomics-based PCR assay for detection of Mycoplasma pneumoniae in a large outbreak in New York State. J Clin Microbiol. 2001;39:1385-90.
- [Google Scholar]
- Detection of viral, Chlamydia pneumoniae and Mycoplasma pneumoniae infections in exacerbations of asthma in children. J Clin Virol. 1999;13:131-9.
- [Google Scholar]
- Molecular detection of Mycoplasma pneumoniae in adults with community-acquired pneumonia requiring hospitalization. J Clin Microbiol. 2001;39:1184-6.
- [Google Scholar]
- Detection of Mycoplasma pneumoniae in spiked clinical samples by nucleic acid sequence-based amplification. J Clin Microbiol. 2002;40:1339-45.
- [Google Scholar]
- Comparison of PCR, culture, and serological tests for diagnosis of Mycoplasma pneumoniae respiratory tract infection in children. J Clin Microbiol. 1999;37:14-7.
- [Google Scholar]
- Rapid, sensitive detection of Mycoplasma pneumoniae in simulated clinical specimens by DNA amplification. J Clin Microbiol. 1992;30:3280-3.
- [Google Scholar]
- , Community-acquired pneumonia in children due to Mycoplasma pneumoniae: Diagnostic performance of a seminested 283 16S rDNA-PCR. Diagn Microbiol Infect Dis. 2001;39:15-9.
- [Google Scholar]
- Practice guidelines for the management of community-acquired pneumonia in adults. Infectious Diseases Society of America. Clin Infect Dis. 2000;31:347-82.
- [Google Scholar]
- Serologic diagnosis of Mycoplasma pneumoniae infections. Enferm Infecc Microbiol Clin. 2006;24(Suppl 1):19-23.
- [Google Scholar]
- Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniae in acute respiratory tract infections in pediatric patients. J Infect Dis. 1996;173:1445-52.
- [Google Scholar]
- Association of Mycoplasma pneumoniae and asthma among Indian children. FEMS Immunol Med Microbiol. 2009;56:25-31.
- [Google Scholar]
- Analysis of children with chlamydophila (Chlamydia) pneumoniae and Mycoplasma pneumoniae respiratoryinfections by real-time PCR assay and serological tests. APMIS. 2008;116:477-83.
- [Google Scholar]
- Sample type is crucial to the diagnosis of Mycoplasma pneumoniae pneumonia by PCR. J Med Microbiol. 2005;54:287-91.
- [Google Scholar]
- Comparison of P1 and 16S rRNA genes for detection of Mycoplasma pneumoniae by nested PCR in adults in Zhejiang, China. J Infect Dev Ctries. 2015;9:244-53.
- [Google Scholar]
- Comparison and evaluation of real-time PCR, real-time nucleic acid sequence-based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae. J Clin Microbiol. 2003;41:4366-71.
- [Google Scholar]





