Translate this page into:
Conference Report on Antimicrobial resistance (AMR) Research Priorities & Action Plan
AmpC and ESBL producing clinical isolates of Klebsiella pneumoniae – A cross-sectional study in a tertiary care teaching hospital, Sikkim
Naresh Chettri1 https://orcid.org/0009-0005-2757-8536, Luna Adhikari1
1Department of Microbiology, Sikkim Manipal Institute of Medical Sciences, Gangtok, Sikkim, India.
E-mail: nareshchettri15@gmail.com
Introduction: Klebsiella pneumoniae causes various types of nosocomial and community-acquired infections. Multidrug resistance (MDR) is seen in Klebsiella spp., which serves as the most common cause of increased morbidity and mortality. Further, the emergence of extended-spectrum beta-lactamases (ESBLs) and AmpC beta-lactamases poses newer diagnostic and therapeutic challenges. This study aimed to know the antibiotic susceptibility profile of K. pneumoniae and production of ESBL and AmpC among these isolates.
Material and Methods: It is a cross-sectional study conducted in the Department of Microbiology, Sikkim Manipal Institute of Medical Sciences, from April 2023 to September 2023. All K. pneumoniae isolates identified by the VITEK 2 Compact system (bioMérieux) from various clinical specimens are included in the study. Antimicrobial susceptibility testing was performed by the VITEK 2 Compact (bioMérieux) system according to the Clinical and Laboratory Standards Institute (CLSI) guidelines 2022. ESBL detection was done by disk diffusion method as per CLSI guidelines 2022. AmpC detection was done by disk diffusion method using cefoxitin and phenylboronic acid.
Results: A total of 72 isolates identified as K. pneumoniae were included in the study. The majority of the isolates were obtained from urine (55.5%), followed by sputum (18%). Aspartate aminotransferase profile of these isolates showed high susceptibility to tigecycline (75%), amikacin (72.2%), and meropenem (69.4%), whereas high resistance to cefuroxime (62.5%) and ceftriaxone (59.7%). Out of 72 strains, 46 strains showed resistance to 2nd as well as 3rd-generation cephalosporins, which is used as an indication for ESBL and AmpC production. Out of 46 isolates tested, ESBL production was seen in 29 isolates (63%) and AmpC production in 15 isolates (32.6%). 37.5% of the total isolates were MDR.
Conclusions: Most of the K. pneumoniae isolates demonstrated resistance to a wide range of antibiotics and high prevalence of beta-lactamases. The findings also highlight the necessity to identify MDR beta-lactamase strains for effective therapy in severe as well as mild infections.
Keywords: AmpC, Extended-spectrum beta-lactamases, Klebsiella pneumoniae, Beta-lactamase
Prevalence of antimicrobial-resistant pathogens and antimicrobial-resistant genes in hospital-generated effluents
Charu Sharma1 https://orcid.org/0009-0001-4206-4905 , Khushbu Batra1, Simran Kandari1, Vivek Kumar1 https://orcid.org/0000-0002-0274-5212
1Himalayan School of Biosciences, Swami Rama Himalayan University, Jolly Grant, Dehradun.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR), in simpler terms, can be referred to as a complex process that gives bacteria the ability to adapt and survive in the presence of inhibitory concentrations of antimicrobial compounds. This process poses a situation of grave concern, evidently from the emergence of pathogens that are resistant to traditionally monitored antimicrobials. Although the evolution of resistant genes is a natural process, the extensive use of antimicrobials has logarithmically accelerated the prevalence of these genes all over the world. Antimicrobials such as antifungals and antibiotics are used in human treatment as well as in the fields of veterinary medicine, animal farming, agriculture, and pisciculture. These antimicrobial agents easily reach surface and groundwater bodies through several routes, such as effluents from wastewater treatment plants, overland flows, and permeation of agricultural water. The sewage treatment plants can be used to extract significant epidemiological data about antimicrobial-resistant pathogens and genes. Furthermore, effluents generated from healthcare facilities act as provenance for the origination of antibiotic-resistant pathogens owing to the presence of a heavy load of patient-associated microbes. The current situation highlights the risk of dissemination of antimicrobial-resistant pathogens and ARGs into the environment from hospital-generated effluents. The World Health Organization has been emphasizing the seriousness of the matter and, in the aftermath of the COVID pandemic, has been urging countries to consider it one of the most imminent threats to humankind. The emergence of new cases of AMR throughout the world is driving researchers to find ways to develop an understanding of the current burden as well as the detection of novel-resistant pathogens. This review is an attempt to highlight the prevalence of various antimicrobial-resistant pathogens and antimicrobial-resistant genes in the influents and effluents generated from health-care facilities. An efficient and sensitive monitoring system can positively influence policymaking and its implementation.
Keywords: Antimicrobial resistance, hospital effluents, antimicrobial-resistant genes, antimicrobial-resistant pathogens
Phytochemicals as potential substitute to antibiotics in opposing microbial drug resistance
Charu Sharma1 https://orcid.org/0009-0001-4206-4905 , Simran Kandari1, Khushbu Batra1, Vivek Kumar1 https://orcid.org/0000-0002-0274-5212
1Himalayan School of Biosciences, Swami Rama Himalayan University, Jolly Grant, Dehradun.
E-mail: nareshchettri15@gmail.com
Phytochemicals, compounds found in plants, have garnered significant attention in recent years as potential substitutes or supplements to antibiotics in combating bacterial drug resistance. Phytochemicals encompass a wide array of compounds, including flavonoids, alkaloids, terpenoids, phenolic acids, and polyphenols, among others. This diversity offers a broad spectrum of bioactive compounds with varying mechanisms of action against bacteria. Many phytochemicals possess inherent antimicrobial properties, which can inhibit the growth of bacteria or even kill them. For instance, flavonoids exhibit antibacterial effects by disrupting bacterial cell membranes or inhibiting essential enzymes. The vast diversity of plant species offers an extensive source of phytochemicals, presenting numerous opportunities for the discovery of novel antimicrobial compounds. This diversity can help overcome the limitations associated with the development of resistance to conventional antibiotics. Nevertheless, antimicrobials from plant-based sources offer enormous potential to tackle bacterial illnesses with no known negative effects. Plant active metabolites include quinines, alkaloids, lectins, flavones, flavonoids, flavonols, coumarin, terpenoids, essential oils, and tannins. While phytochemicals hold promise as alternatives or complement to antibiotics, several challenges remain, including standardization of extracts, understanding their mechanisms of action, and ensuring their safety and efficacy through rigorous scientific investigation.
Keywords: Antimicrobial drug resistance, plant metabolites, bioactive compounds
Isolation and molecular characterization of the novel mycobacteriophage KRDG1 and its specificity determinants
Anuja Kakkar1 https://orcid.org/0000-0002-9152-3741 , Tanmayee Nayak2, Garima Kandwal2, Lav Kumar Jaiswal2, Ankush Gupta2
1Department of Biochemistry, Interdisciplinary School of Life Sciences1, Institute of Science, Banaras Hindu University, 2Department of Biochemistry, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India.
E-mail: nareshchettri15@gmail.com
Tuberculosis represents a substantial global health challenge. Antibiotics have historically proven effective, but the increasing problem of antibiotic resistance compels us to investigate alternative strategies. Alternative therapies such as phage therapy and vaccines have been introduced, especially considering the limited discovery of new antibiotics. Phages exhibit the ability to selectively attach to specific bacteria, making it a targeted approach for eliminating bacterial infections without disrupting the normal microbial balance. Bacteriophages that infect the genus Mycobacterium are commonly referred to as mycobacteriophages. To isolate phages, Mycobacterium smegmatis mc2155 is the most frequently used non-pathogenic host. For clinical potential, screening includes pathogenic Mycobacterium tuberculosis H37Rv. Host specificity relies on receptor binding proteins (RBPs) on phage tails, comprising tail fibers and tail spike proteins (TSPs). Typically, TSPs are short spikes with enzymatic activity, while tail fibers are lengthy fibrous proteins that exclusively bind to receptors as they lack enzymatic activity. A new mycobacteriophage named KRDG1 was isolated from Ravidas Ghat in Varanasi, falling within the K1 cluster and belonging to the Siphoviridae family. Its genome is 58681 bp long, with 96 genes, including 1 tRNA; 39 genes are functionally annotated, while 56 are hypothetical proteins. Five genes were identified as potential (RBPs), and gene gp26 (80 kDa) was shown to display enzymatic activity against M. smegmatis mc2 155, indicating its role as a potential RBP/TSP. Phage therapy, informed by the knowledge of TSPs, enables the selection of diverse phages for broad antibiotic-resistant strain coverage.
Keywords: Mycobacterium, Antibiotic resistance, Bacteriophages, Receptor binding proteins, Tail spike proteins
A novel bacteriophage, isolated from hospital sewage, effectively mitigates the biofilm of clinical extensively drug-resistant strains of Klebsiella pneumoniae
Sambuddha Chakraborty1 https://orcid.org/0000-0002-4536-7223 , Anusha Rohit2, Ashwini Chauhan1,2 https://orcid.org/0000-0002-0555-8244
1Department of Microbiology, Tripura University, Agartala, Tripura,
2Department of Microbiology, Delhi University, BK, Bachhawat Block, University of Delhi South Campus, New Delhi, India.
E-mail: nareshchettri15@gmail.com
Klebsiella pneumoniae, a member of the ESKAPE pathogen group, presents formidable global treatment challenges due to its severe antibiotic resistance, raising global concerns about its resistance to available antibiotic therapies. Furthermore, in addition to antibiotic resistance, the pathogen’s ability to form biofilms on tissue or device surfaces contributes significantly to poor treatment outcomes by enhancing antibiotic resistance. As the discovery of antibiotics is slower than the emerging resistance, a full-proof anti-biofilm strategy is warranted. Bacteriolytic viruses are emerging as a promising alternative treatment for combating bacterial infections. In this study, we report the isolated and purified lytic bacteriophage of the family Casjensviridae from hospital sewage, which lysed the clinical strains of carbapenem-resistant K. pneumoniae. Further, we found that the virus efficiently reduced the biomass and viable cell count of the preformed biofilm by the multidrug-resistant clinical K. pneumoniae strains. Electron microscopy corroborated our results and showed that the phage could disrupt biofilms, lysing bacterial cells underneath the extracellular polymeric substances. The crystal violet assay indicates maximum biofilm reduction after 12 h of treatment. Bacterial colony count also confirmed the effectiveness of phage treatment in reducing K. pneumoniae biofilm. With drug resistance on the rise, there is a pressing demand for safe and efficient treatment alternatives; phages, the natural predators of bacteria armed with bacteriolytic properties, offer promise. Extensive studies confirm phage safety, and their evolutionary adaptability makes them indispensable against ever-changing bacteria.
Keywords: Antimicrobial resistance, Klebsiella pneumoniae, Bacteriophage, Alternative to antibiotics
Systems, methods, and devices to detect drug resistance: BL testers
Niteesh Kumar Pandey1 https://orcid.org/0000-0002-4919-5203 , Saugata Hazra1 https://orcid.org/0000-0002-3074-1534
1Department of Bioscience and Bioengineering, Indian Institute of Technology-Roorkee, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com; saugata.iitk@gmail.com
Hazra-Lab developed three types of Beta-lactamase (BL) Testers for on-site detection of drug-resistant bacteria: (1) BL-tester Basic, (2) BL-tester Advance, and (3) BL-tester Environmental. BL-tester Basic has three components: Sample preparation vial, BL-tester vial, and dye-containing pouch. Antibiotics were used to screen-resistant bacteria in the BL-tester basic. It can work with milk, urine, and soil samples. BL-tester Advance would be based on bacteria culturing to reduce the detection time, i.e., about 2 h. It can work with clinical samples such as body fluids (saliva, pus, and blood). BL-tester Environmental could detect drug-resistant bacterial presence within 30 min. This system can work to detect drug-resistant bacteria in wastewater, river water, ponds, and sewage samples. BL Environmental works on four types of filter chambers, and filter size could be from 1 mm to 0.45 μm diameter to concentrate bacteria. Chromogenic dye has been used in every BL Tester, which can change its color from yellow (λmax 390) to red (λmax 486) in the presence of drug-resistant bacteria. The total duration to detect drug resistance was 5 h for BL tester Basic, while BL tester advance took 2 h and Environmental took only 30 min. The naked eye visualizes results and can be analyzed by an untrained person. The intensity of the color (red) was directly proportional to drug-resistant bacterial load in the samples. BL Testers are easy, fast, reliable, and less costly for detecting drug-resistant bacteria in clinical and non-clinical samples.
Keywords: Antimicrobial resistance, Quick detection, Chromogenic, Devices
Comprehensive analysis of tuberculosis dynamics: A comprehensive analysis of 1-year secondary data from the DTO office in Dehradun
Anil Kumar Bisht1 https://orcid.org/0009-0009-8354-2756 , Himanshu Mamgain1, Abhay Srivastava1
1Department of Community Medicine, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Walakhur, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: This study presents a holistic examination of tuberculosis dynamics through an in-depth analysis of various key factors, including treatment outcomes, sociodemographic characteristics, and the prevalence of co-morbidities in the Dehradun region.
Objectives: (i) Conduct a comprehensive analysis of tuberculosis dynamics, encompassing treatment outcomes, sociodemographic profiles, and the prevalence of diabetes and human immunodeficiency virus (HIV) among cases. (ii) Examine the distribution of tuberculosis cases based on types and sites, along with an analysis of programmatic management of drug resistant tuberculosis (PMDT) regimen types employed in management.
Methodology: Utilizing secondary data, the study employs rigorous statistical methods to analyze tables related to treatment outcomes, sociodemographic, diabetes, HIV status, types of cases, PMDT regimen types, and sites of disease.
Results: This study provides a comprehensive analysis of tuberculosis cases based on secondary data obtained from the District Tuberculosis Office in Dehradun. Key findings reveal significant patterns in age distribution, with a substantial proportion in the 21–30 age group. Males constitute the majority of cases (58.4%), while females account for 41.5%. Diabetes status indicates that 8.2% of cases are diabetic, with 90.1% non-diabetic. HIV prevalence is low, with 94.9% non-reactive cases. New cases dominate (90.2%), and most cases exhibit a non-resistant PMDT regimen (97.0%). Pulmonary cases prevail at 68.1%, and the majority exhibit treatment completion (39.5%). Notably, a considerable percentage remains unresponsive across various categories. These findings offer crucial insights for targeted interventions and public health strategies in managing tuberculosis in the Dehradun region.
Implications: The study’s insights hold implications as follows: (i) Tailoring treatment strategies for improved outcomes, (ii) addressing sociodemographic disparities in tuberculosis prevention and management, and (iii) informing targeted interventions for tuberculosis patients with comorbidities.
Conclusion: This research contributes valuable knowledge to the understanding of tuberculosis dynamics, facilitating the development of targeted and effective public health interventions in the Dehradun region.
Keywords: Tuberculosis dynamics, Secondary data analysis, PMDT regimen types
Emerging trends in multidrug-resistant mechanisms and therapeutic strategies: A comprehensive study
Abha Verma1, Monika Bajpai2, Sarita Singh1 https://orcid.org/0000-0002-9671-4539
1Department of Microbiology, School of Life Science and Technology, IIMT University, Meerut, 2Department of Microbiology, School of Life Science and Technology, IIMT University, Meerut, Uttar Pradesh, India.
E-mail: nareshchettri15@gmail.com
This review provides a comprehensive exploration of the dynamic landscape of multidrug resistance (MDR), a critical challenge in the context of infectious diseases. Beginning with an insightful introduction, the review defines MDR and underscores its significance in public health. Historical perspectives and key milestones in MDR research are presented, offering a contextual foundation for understanding the evolution of drug resistance. The molecular intricacies underlying MDR are also elucidated in the next section, delving into the genetic and molecular mechanisms that drive resistance. Specific genes, proteins, and pathways implicated in MDR are highlighted, providing a detailed understanding of the molecular underpinnings of this complex phenomenon. The review also examines the evolution of drug resistance in pathogens and explores how microorganisms develop and adapt to resist drugs. The role of selective pressure and adaptation is scrutinized, shedding light on the dynamic interplay between pathogens and therapeutic agents. Further, it reviews state-of-the-art technologies and methodologies employed in MDR research. From omics approaches to structural biology and computational modeling, this analysis showcases the innovative tools that have revolutionized our understanding of MDR mechanisms. Addressing the current challenges and opportunities in combating MDR, the review also navigates through the obstacles faced in clinical settings. Ongoing research and potential breakthroughs are discussed, providing a glimpse into the future of MDR management. The review provides existing and emerging therapeutic strategies for overcoming MDR. Potential avenues for future research and intervention are explored, providing a roadmap for the development of effective countermeasures against drug-resistant pathogens. Finally, it analyzes the clinical implications of MDR on outcomes and public health. Strategies for prevention and control are explored, emphasizing the importance of a holistic approach to address the significant impact of MDR on global health. This comprehensive study not only consolidates existing knowledge but also propels our understanding of MDR mechanisms and therapeutic strategies, offering valuable insights for researchers, clinicians, and policymakers in the ongoing battle against antimicrobial resistance.
Keywords: Multi-drug resistant, pathogen, microorganism
Revolutionizing antibiotic-resistant detection: A breakthrough paper-based approach
Seema Nautiyal1, Pratyush Kumar Sahoo1, Nikku Yadav1 https://orcid.org/%25200000-0002-0315-4895
1Department of Clinical Research, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The abstract outlines a groundbreaking paper-based approach designed to tackle the global challenge of antimicrobial resistance (AMR) by detecting antibiotic resistance in superbugs, which are bacteria resistant to multiple antibiotics. These superbugs pose a formidable obstacle in the effective treatment of infections. The innovation discussed in the abstract could have far-reaching implications for the rapid identification of antibiotic-resistant bacteria, guiding treatment decisions, and curbing the spread of drug-resistant infections. The context provided highlights the severity of the AMR issue, particularly in India, where 297,000 deaths in 2019 were attributed to AMR, with 1,042,500 deaths associated with the problem. Researchers at the Indian Institute of Science and Jawaharlal Nehru Centre for Advanced Scientific Research in Karnataka devised a strategy incorporating biphenyl-4-carboxylic acid (BCA) within a supramolecular hydrogel matrix containing Terbium Cholate (TbCh). This hydrogel exhibits green fluorescence under ultraviolet light. The scientists developed an enzyme substrate by linking BCA to the cyclic beta-lactam ring, a component of the antibiotic. In the absence of the beta-lactamase enzyme, responsible for breaking down the antibiotic, the TbCh hydrogel remains non-fluorescent. However, in the presence of the enzyme, the gel emits a green fluorescence. This luminescence serves as an indicator of the presence of antibiotic-resistant bacteria, with the intensity correlating with the bacterial load. Non-resistant bacteria, in contrast, display significantly lower green intensity, facilitating their differentiation from resistant strains. The proposed method aims to offer an efficient tool for the detection and monitoring of antibiotic resistance, potentially revolutionizing treatment strategies against superbugs.
Keywords: Superbugs, BCA, Fluorescence, Beta-lactamase, Hydrogel Matrix
Advancing antimicrobial therapy through nanotechnology: Design, evaluation, and clinical translation
Sandeep Rawat1, Vedika Rawat1, Nikku Yadav1 https://orcid.org/%25200000-0002-0315-4895
1Department of Clinical Research, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance presents a formidable challenge to global public health, necessitating innovative therapeutic strategies. Nanotechnology-based approaches offer promising solutions by harnessing the unique properties of nanomaterials for targeted antimicrobial therapy. This abstract provides a comprehensive overview of the research landscape surrounding nanotechnology-based antimicrobial therapy, focusing on design, evaluation, and clinical translation. In the design phase, meticulous selection and engineering of non-materials are paramount to optimize their physicochemical properties for enhanced antimicrobial efficacy. Surface functionalization techniques enable precise control over drug loading and release kinetics, facilitating targeted delivery and reducing off-target effects. Moreover, the incorporation of antimicrobial agents within nanoparticles enhances drug stability and bioavailability while minimizing the emergence of resistance. Evaluation of nanotherapeutics begins with rigorous in vitro studies to assess antimicrobial activity against a broad spectrum of pathogens, including multidrug-resistant strains. Mechanistic investigations elucidate the underlying modes of action, providing insights into nanoparticle-mediated antimicrobial effects. Subsequent in vivo studies in animal models evaluate pharmacokinetics, biodistribution, and safety profiles, laying the groundwork for clinical translation. Clinical translation of nanotechnology-based antimicrobial therapies involves navigating regulatory pathways and conducting systematic clinical trials. Ongoing research endeavors aim to address critical challenges such as nanotoxicity, scalability, and long-term efficacy. In conclusion, nanotechnology holds tremendous promise in revolutionizing antimicrobial therapy, offering targeted and efficient interventions against antimicrobial-resistant pathogens. Continued research efforts are essential to overcome existing barriers and realize the full potential of nanotechnology in combating infectious diseases.
Keywords: Nanotchnology, physiological, targeted delivery, nanotherapeutics, nanotoxicity
Anti-microbial resistance: A global threat to humanity
Anjali Mishra1, Ruchika Thapliyal1, Nikku Yadav1
1Department of Clinical Research, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) refers to microorganisms’ ability to resist the effects of antimicrobial drugs, rendering previously effective treatments ineffective. AMR poses a significant threat, leading to infections that are challenging or impossible to treat. This results in prolonged illness, increased mortality rates, and higher health care. The World Health Organization 2014 Global Report on AMR highlighted gaps in surveillance networks worldwide. The 2016 UN General Assembly emphasized AMR’s importance and urged countries to commit to the National Action Plan. In 2019, drug-resistant infections caused 4.95 million global deaths, with 1.27 million directly attributed to AMR. Low- and middle-income countries bear a disproportionate burden. Without intervention, AMR-related deaths could escalate to 10 million annually by 2050 [Table 1]. AMR is a critical global challenge affecting public health and development. In 2019, bacterial AMR caused 1.27 million global deaths, contributing to a staggering 4.95 million deaths overall. Misuse of antimicrobials drives drug-resistant pathogens. AMR transcends borders, impacting countries across income levels. It jeopardizes modern medicine, making infections harder to treat. The antibiotic pipeline crisis demands research and equitable access to vaccines and diagnostics. The World Bank estimates US$ 1 trillion in additional healthcare costs by 2050 and annual gross domestic product losses of US$ 1 trillion to US$ 3.4 trillion by 2030. AMR poses a significant public health threat in India. In 2019, 297,000 deaths attributable to AMR were reported, and 1,042,500 deaths associated with AMR were reported, and India ranks 145th globally in age-standardized mortality rate related to AMR. The tabulated numerical data mentioned below highlight the scale and urgency of the AMR crisis and the need for comprehensive global interventions.
Keywords: AMR, global crises, global economy, public health burden, national action plan
| Aspect of AMR | Data related to AMR globally. |
|---|---|
| Estimated deaths due to AMR annually | Over 700,000 |
| Economic burden of AMR annually | Approximately $1 trillion globally |
| Percentage of antibiotics misused | Around 30% in humans, up to 80% in animal agriculture |
| Estimated increase in deaths by 2050 | Up to 10 million annually if no action is taken to address AMR |
| Resistance rates in common bacteria | Vary widely by region and antibiotic, ranging from 20% to over 70% |
| Investment needed for R and D | Estimated at least $2 billion annually to develop new antimicrobial agents |
| Estimated cost savings from AMR interventions | Up to $100 billion annually through reduced healthcare costs and productivity losses |
| Number of countries with national action plans on AMR | Over 150 countries have developed national action plans to address AMR |
AMR: Antimicrobial resistance
Clinical application of bacteriophages to treat multidrug resistance: Phage journey from bench to bedside
Akankska Rawat1, Swati Pandey1, Nikku Yadav1 https://orcid.org/0000-0002-0315-4895
1Department of Clinical Research, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Bacteriophages (phages) are viruses that infect and kill specific bacteria. They offer an alternative to antibiotics for treating infections caused by multidrug-resistant (MDR) bacteria, which are a major threat to global public health. Phages have different mechanisms of action from antibiotics, such as lysing the bacterial cell wall, and can overcome some of the limitations of antibiotics, such as resistance development, toxicity, and biofilm formation. However, phage therapy also faces some challenges, such as phage specificity, bacterial immunity, regulatory approval, and clinical trial design. Phage therapy has been used in some countries for decades, but its clinical application is still limited to specific cases of patients with few or no other options. There have been some recent studies that demonstrate the efficacy and safety of phage therapy in animal models and human patients, especially in combination with antibiotics. The phenomenon of phage-antibiotic synergy, in which antibiotics enhance phage production or phages increase antibiotic susceptibility, has been observed and explored. Phage therapy has shown promising results in treating MDR infections caused by bacteria such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Staphylococcus aureus. Phage therapy is a potential solution for the growing problem of MDR bacteria, but it requires more research and development to optimize its clinical application. Phage-antibiotic synergy is based on various mechanisms that enhance bacterial killing. Some phages and antibiotics may also show antagonism or indifference. More research is needed to understand the interactions between phages and antibiotics in vitro and in vivo. Phage-antibiotic combinations have shown promising results in some animal and human infections, especially with MDR bacteria. Clinical trials should determine the optimal conditions and efficacy of phage-antibiotic therapy.
Keywords: Multi drug resistant, phage therapy, biofilm, phage production, bacteriophages
Isolation and prevalence of urinary tract infection causing organism and their drug-resistant pattern in Haridwar, Uttarakhand
Sumit Chand1 https://orcid.org/0009-0002-4538-9585 , Annapurna Katara1, Vijeta Chaudhry1, Sagar Vishwakarma1, Harish Chandra1
1Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar, Uttrakhand, India.
E-mail: nareshchettri15@gmail.com
One of the main and most frequent infections in women is urinary tract infections (UTIs), and it has been reported that nearly all women in their lives have experienced UTIs at least once. Escherichia coli is the most frequent causative microbe for UTIs, and the most concerning aspect of UTI isolates is their resistance to conventional antibiotics. A number of factors, including irrational and excessive use of antibiotics in clinical settings, animal husbandry, and agriculture, may have contributed to the emergence of multi-drug resistant, extensively drug-resistant, and biofilm-forming microorganisms, as well as accelerated the process of antimicrobial drug resistance manyfold. Nowadays, the pace at which antibiotic resistance is being acquired is far higher than the rate at which new chemically synthesized drugs are designed and developed. The population could succumb as a result of this delay, as the COVID-19 pandemic of 2019 has shown. One-fourth of all infections connected to health care worldwide are UTIs, which are among the most despised infectious diseases. For UTI patients, the infection may be emotionally and financially devastating, in addition to being unbearably uncomfortable. A wide spectrum of microbes, such as E. coli from the normal microflora or other Gram-positive or Gram-negative bacteria from the surrounding environment, may have colonized and caused the illness. E. coli is the pathogen that is most commonly linked to UTIs, followed by various Proteus species and members of the Klebsiella species. Given the increased acquisition of extended-spectrum β-lactamases by members of the Enterobacteriaceae family, multidrug resistance in uropathogenic organisms poses a chronic and unrelenting danger to public health. We are concentrating on the incidence of UTI and drug resistance in the current research on the significance of drug resistance in uropathogens and the severity associated with the disease. The study entails surveying the district’s (Haridwar) general populace to get information on the prevalence of the causal organisms and the efficacy of the different antibiotics that are administered to treat them.
Keywords: Multi-drug resistant, Extensive drug-resistant, Uropathogens, Extended-spectrum β-lactamases, Urinary tract infections
Detection methods for antimicrobial resistance: Phenotypic and genotypic
Rohit Pawar1, Nikku Yadav1 https://orcid.org/0000-0002-0315-4895
1Department of Clinical Research, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Screening methods for antimicrobial resistance (AMR) are crucial for effective management of infections and surveillance of resistance patterns. There are some phenotypic methods that directly assess the ability of microorganisms to resist antimicrobial agents. Disk diffusion, broth microdilution, and E-test are common phenotypic techniques that determine resistance based on growth inhibition. Genotypic methods, on the other hand, detect specific genetic markers associated with resistance. Polymerase chain reaction, DNA sequencing, and hybridization techniques identify resistance genes or mutations. These methods are valuable for rapid and accurate identification of resistance mechanisms. Emerging technologies such as matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry and whole-genome sequencing (WGS) offer advanced screening capabilities. MALDI-TOF can rapidly identify microbial species and detect resistance markers. WGS provides a comprehensive analysis of microbial genomes, aiding in the identification of novel resistance mechanisms. Choosing the appropriate screening method depends on various factors, including the type of microorganism, the antimicrobial agent of interest, and the purpose of screening. Combinations of phenotypic and genotypic methods are often used to enhance sensitivity and specificity. Overall, screening methods play a critical role in guiding treatment decisions, designing effective infection control measures, and monitoring AMR trends at local and global levels.
Keywords: Genotypic, phenotypic, genome, hybridization, MALDI-TOF
Overexpression of PDR16 confers amphotericin B resistance in plasma membrane proteolipid 3-dependent manner
Sapna Kalra1, Sunita1, Vinay Kumar Bari1 https://orcid.org/0000-0002-5670-3798
1Department of Biochemistry, Central University of Punjab, Bathinda, Punjab, India.
E-mail: nareshchettri15@gmail.com
Invasive fungal infections are the major cause of morbidity and mortality in immunosuppressed patients, which are becoming more widely acknowledged. A common polyene antifungal drug, amphotericin B (AmB), binds to plasma membrane ergosterol and causes cellular ions to leak, which results in cell death. PDR16, a gene for pleiotropic drug resistance in Saccharomyces cerevisiae, is extremely resistant to AmB when expressed in multicopy. However, the mechanism of PDR16-mediated AmB resistance is not clear. Here, we provided evidence that a plasma membrane proteolipid 3 protein encoded by PMP3 plays a crucial role in PDR16-mediated AmB resistance. In addition, ablation of the sphingolipid biosynthesis genes FEN1, SUR4, and regulatory gene YPK1 inhibited PDR16-mediated AmB resistance, indicating that this pathway is involved in PDR16-mediated AmB resistance. Moreover, we revealed that pmp3∆ deletion reduced membrane integrity and cellular phytosphingosine content, as well as enhanced AmB binding ability in PDR16 overexpressing cells, resulting in enhanced AmB sensitivity.
Keywords: Fungal infection, amphotericin, resistance
Virtual screening and molecular docking to identify the sterol transport protein osh4-inhibiting drugs
Sunita Tanwar1, Sapna Kalara1, Tanu Singh1, Vinay Kumar Bari1 https://orcid.org/0000-0002-5670-3798
1Department of Biochemistry, Central University of Punjab, Bathinda, Punjab, India.
E-mail: nareshchettri15@gmail.com
Oxysterol-binding proteins mainly involved in the transport of sterol and phosphatidylinositol 4-phosphate between membranes such as plasma membrane and endoplasmic reticulum are highly conserved from yeast to humans. Ergosterol, the main constituent of the yeast plasma membrane, influences sterol-targeting drug action, such as azoles and polyenes, allylamine, and morpholine. Amphotericin B (AmB) is a polyene drug commonly used to treat fungal infection caused by pathogenic fungi Candida albicans and Cryptococcus neoformans. Recent studies have demonstrated that sphingolipid biosynthesis defective mutants (FEN1, SUR4, and PMP3) also play a critical role in AmB drug-resistant modulation, but the exact mechanism involved is still unclear. Pathogenic fungi have been described as a global health threat due to their ability to cause invasive infections with a high mortality rate. The emergence of antifungal drug resistance in pathogenic fungi prompts us for novel drug discoveries which are required urgently for combating pathogenicity caused by these pathogenic fungi. The current study was intended to identify osh4 inhibitors with a high binding affinity. Structure-based virtual screening was carried out on approved medications against osh4 with the help of AutoDock VINA, which is included in the PyRx 0.8 package. The compound with the highest affinity was then examined, and structurally comparable compounds were docked to the osh4 protein once again to discover a new and more effective inhibitor molecule against osh4. This study will provide mechanistic insight into the role of sphingolipids and oxysterol binding protein in antifungal drug resistance that could be used to develop novel drug formulations against life-threatening diseases caused by fungal pathogens.
Keywords: Yeast, amphotericin, multi-drug resistance
Using a discrete renewal process to assess the impact of non-pharmaceutical interventions and the role of severe acute respiratory syndrome coronavirus 2 virus mutations in disease transmission and mortality among Indian population
Meghna Banerjee1 https://orcid.org/0009-0003-8540-1988 , Prithwish Ghosh2, Arindom Chakraborty2
1Department of Bio-science and Biotechnology, Banasthali Vidyapith, Tonk, Rajasthan, 2Department of Statistics, Visva-Bharati University, Santiniketan, Bolpur, West Bengal, India.
E-mail: nareshchettri15@gmail.com
In recent years, the world has not suffered so much compared to the devastation caused by COVID-19. This study aims to identify, among the myriad of mutations gained by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome throughout the first and second waves in India, and those novel mutations which, despite the interventions imposed by the Indian Government, have contributed to virulence, drug resistance, vaccine evasion, and resistance to the antibodies produced by the host immune system. We consider a stochastic model based on a discrete renewal process that includes various controlling measures to systematically evaluate their effects on the disease transmission dynamics through three interlinked components. A Bayesian model has been considered for the infection cycle to observe deaths with upper and lower bounds of the total population infected (attack rates), case detection probabilities, and the reproduction number over time. The MCMC technique was adopted to analyze the data. In this study, we treat interventions as covariates in modeling the average reproduction number. Here, the time-varying reproduction number (Rt) has been assumed to be a piece-wise constant function that starts from a baseline prior, and mutations are used as covariates along with the nonpharmaceutical interventions. Thus, this study will aid immensely in developing genomic surveillance strategies for monitoring SARS-CoV-2 mutants and potential approaches against antimicrobial resistance in the future.
Keywords: Severe acute respiratory syndrome coronavirus 2, Single nucleotide polymorphism, Non-pharmaceutical interventions, Antimicrobial resistance, Time-varying reproduction number (Rt), Genomic surveillance
Advancement in alternative strategies to combat antimicrobial resistance
Rohit Bhardwaj1 https://orcid.org/0000-0002-9933-0864 , Satyendra Kumar Rajput1
1Department of Pharmaceutical Sciences, Gurukula Kangri Deemed to be University, Haridwar, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) poses an escalating global threat to public health, demanding innovative strategies to combat the rising tide of resistant pathogens. This study explores the latest disruptive approaches in the fight against AMR, focusing on the development and application of novel antimicrobials, vaccines, and immunomodulators. The paper delves into cutting-edge research and breakthroughs that promise to reshape the landscape of infectious disease control. The first section of the study examines the evolution of antimicrobial agents, highlighting recent advancements in the discovery and design of potent and selective compounds. Novel antimicrobials, ranging from synthetic molecules to naturally derived alternatives, are explored for their efficacy, safety profiles, and potential to overcome resistance mechanisms. The second segment underscores the pivotal role of vaccines in preventing and mitigating the spread of resistant pathogens. An in-depth analysis of next-generation vaccines, including subunit vaccines, RNA-based platforms, and innovative delivery systems, provides insights into their potential to elicit robust and long-lasting immune responses. The third component focuses on immunomodulators as a promising avenue to enhance the host’s innate and adaptive immune responses. Therapeutic interventions that modulate the immune system, such as cytokine therapies, immune checkpoint inhibitors, and targeted immunotherapies, are evaluated for their capacity to bolster the body’s defense mechanisms against drug-resistant microbes. Furthermore, the study critically assesses the challenges and opportunities associated with the translation of these disruptive approaches from bench to bedside, emphasizing the need for interdisciplinary collaboration, regulatory support, and global cooperation. In conclusion, this review underscores the urgency of adopting a multifaceted and dynamic approach to counter AMR. By embracing novel antimicrobials, advancing vaccine technologies, and harnessing immunomodulatory strategies, the global community stands poised to revolutionize its response to antimicrobial resistance, safeguarding public health for generations to come.
Keywords: Anti-microbial resistance, Anti-microbials, Immunomodulators, Vaccines
Fungal isolates from degraded wood showing mannanase enzymatic hydrolysis activity
Vijeta Chaudhry1 https://orcid.org/0009-0007-9359-3931 , Sagar Vishwakarma1, Sumit Chand1, Harish Chandra1
1Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Enzymes generated from microorganisms are useful in a wide range of biotechnological applications, including bioconversion, biotransformation, and other biological uses. Mannan enzymatic hydrolysis is more selective and environmentally benign than chemical or heat procedures. Filamentous fungus is an excellent source for commercial enzyme synthesis. They may generate and secrete enormous quantities of extracellular enzymes. Mannans and xylans, in addition to cellulose and lignin, are the most abundant carbohydrate components of wood lignocellulosic materials. Mannanases, mannosidases, and glucosidases are the three primary enzymes that break down mannan. To remove side-chain substituents, additional enzymes such as β-galactosidases and acetyl mannan esterase are necessary. Microbial mannanases have found application in pulp and paper, pharmaceutical, food, feed, oil, and textile industries. Based on an understanding of the interactions between many critical components, <AQ69>RSM </AQ69>is used to improve the process parameters for mannanase synthesis. Mannanase is made utilizing expensive pure mannans such as konjac, locust bean gum (LBG), and guar-gum, making the enzyme pricey and difficult to deploy economically. The current work attempts to identify mannan-degrading fungus from a deteriorated wood supply. LBG was used to screen mannanase-producing fungus. The isolates were described on a morphological, physiological, and biochemical basis. These isolates were then tested on a medium using LBG as a mannan source. Physiological characterization was performed using conventional protocols, and the three fungi were identified using an enzyme hydrolysis test. The culture filtrate showed the highest level of activity, according to the study.
Keywords: Mannan, Mannanase, Hydrolysis, Enzymes, Locust bean gum
Antimicrobial activity of fungal endophytes isolated from high-altitudinal medicinal plants
Sagar Vishwakarma1 https://orcid.org/0009-0006-9508-2177 , Vijeta Chaudhry1, Sumit Chand1, Harish Chandra1
1Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Endophytes are microorganisms that reside within the plants and are capable of invading plant tissues without causing damage to the plants and, by residing within their tissues, impart beneficial attributes to them. Endophytes maintain an association with plants throughout a minimum portion of their life cycles. Other organisms besides bacteria and fungi have the potential to function as endophytes. The utilization of these endophytes may prove beneficial in the exploration of bioactive compounds with potential pharmaceutical and medical uses. Fungi are recognized as a significant source of natural products and produce secondary metabolites in abundance. The objective of this study was to assess the antimicrobial activity of ethyl acetate extracts derived from the fungus Aspergillus flavus, which were obtained from the leaves of the plant Roscoea purpurea. The extract was obtained by inoculating potato dextrose broth medium with A. flavus and subsequently extracting filtrates of both fungi with ethyl acetate 1:1 (v/v) and evaporating. The extract of fungus was evaluated for its antimicrobial activity against some Gram-positive (Staphylococcus aureus, Bacillus subtilis, Bacillus cereus, and Streptococcus pneumoniae) and Gram-negative bacteria (Escherichia coli, Salmonella typhi, Klebsiella pneumoniae, and Pseudomonas aeruginosa). Results demonstrated that the ethyl acetate extract of the fungus possessed potent antibacterial properties. Using the agar diffusion method, the minimum zone of inhibition against B. subtilis (19.5 ± 0.56 mm) and the maximum zone of inhibition against S. pneumoniae (30.75 ± 0.49 mm) were observed. These results suggest that endophytic fungi originating from R. purpurea plants could serve as viable sources of antimicrobial agents and find application in the pharmaceutical industry.
Keywords: Endophyte, Fungi, Antimicrobial agent, Pharmaceutical industry
Epiphytic fungi associated with seaweeds, their economical importance, and future prospects
Jatin Kumar1 https://orcid.org/0009-0007-1769-3017 , Felix Bast1
1Department of Botany, Central University of Punjab, Bathinda, Punjab, India.
E-mail: nareshchettri15@gmail.com
The fungus that colonizes the outer surface of the host (seaweed) is known as epiphytic fungi or algicolous fungi, and they are ubiquitous, can be saprophytic, parasitic, or in symbiotic association with its host, and play important roles in benthic communities and almost all of the organisms including macroalgae or terrestrial plants mediate ecological signaling and interactions with nearby microbial community or the fungi associated with it by the emission of low or high molecular weight compounds called infochemicals and when perceived by either host or associated organism can influence growth, defense mechanisms and metabolic pathways of both emitter and receiver, these compounds have different chemical structures, different action ranges and it can act as defense against pathogens and other surrounding holobiont members that compete for the nutrients. The compounds released by seaweeds are mostly sugars, amino acids, and organic acids. In recent years, the interest of many scientists increased significantly in marine fungi due to their ability to produce different novel products that hold significant therapeutic value and cause of their unusual habitat. According to Kohlmeyer, almost 30% of these known marine fungi are associated with seaweed and day by day due to overuse of antibiotics in animal husbandry and livestock farming, and as a result, there was a significant rise in cases of antibiotic resistance in the microbial communities which have lead to the emergence of resistant microbes and superbugs globally as well as endemically and caused scientists to go back to nature and search for novel bioactive compounds which could be potential drug target to fight with the global antimicrobial resistance.
Keywords: Antimicrobial resistance, Infochemicals, Bioactive compounds, Epiphytic fungi
Metagenomic analysis of the Ganga water’s microbial diversity to assess its self-cleaning potential
Annapurna Katara1 https://orcid.org/0009-0007-5999-1274 , Sumit Chand1, Ramesh Chandra Dubey1
1Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar, Uttrakhand, India.
E-mail: nareshchettri15@gmail.com
The Ganga is revered by people for supplying the environment and ecology that are essential to life and its sustenance. It contributes to the region’s increased food security by offering a sizable agricultural area. A wide variety of microorganisms, such as bacteria, viruses, and fungi, are found in the river. Whole genome sequencing and metagenomic analysis of pooled samples drawn from three distinct places along the river’s length are included in our study. Obtained raw sample fastq reads from the Illumina platform were assessed for quality using FastQC. The raw fastq reads after the quality assessment are subject to processing through Fastp and further reassessed for quality using FastQC. For metagenomic classification and meta-taxonomic analysis, the processed paired-end reads will then be uploaded to CCMetagen and MG-RAST online servers, respectively. The OTU tables thus obtained were then used to deduce a phylogenetic tree using NCBI’s common tree file and will be plotted by using an interactive tree of life. This study brings forth the alpha diversity of the water sample under study and enables us to categorize it according to its relative taxonomic abundances among various levels of the hierarchy, including phyla, classes, orders, families, and genera. The taxonomic richness and evenness at different hierarchical levels were also obtained from the sequenced data. This study brings forward an in-depth analysis of the correlation between microbial diversity and self-cleansing potential of Ganga water.
Keywords: Metagenomics, Ganga water, Taxonomic classification, Microbial diversity
In silico strategies for combatting drug resistance in Mycobacterium tuberculosis: Screening inhibitors targeting GlfT2 enzyme
Ankit Verma1, Vijay Kumar1 https://orcid.org/0000-0002-9571-561X , Bindu Naik2, Vivek Kumar1, Sanjay Gupta1
1Himalayan School of Biosciences, Swami Rama Himalayan University, Dehradun, India, 2Department of Food Science and Technology, Graphic Era Deemed to be University, Clement Town, Bell Road, Dehradun, UK, India-248002
E-mail: nareshchettri15@gmail.com
The World Health Organization designates tuberculosis (TB) as a global health crisis, causing 1.5 million deaths annually. Efforts to control TB globally face challenges due to traditional drug development methods lacking a structural-based approach, hindering the production of broad-spectrum drugs. Arabinogalactan (AG), a fundamental component of bacterial cell walls, has emerged as a promising target for pharmaceutical intervention. Inhibition of the GlfT2 enzyme, critical for AG synthesis, holds the potential to thwart mycobacterial cell formation. To identify potential candidates, a virtual screening of 30,417 compounds from three repositories was conducted using AutoDock Vina, with GlfT2 as the protein target. Compounds exhibiting the highest binding affinity and inhibition constant (KI) were then subjected to assessments of drug-likeness, pharmacological characteristics, and molecular dynamics simulation (MDS). Results revealed that compounds CSID54154 (−10.7 Kcal/mol; KI 475.24 pM), CSID67239 (−9.2 Kcal/mol; KI 138.09 uM), DB12983 (−13.5 Kcal/mol; KI 1.05 nM), DB12424 (−12.8 Kcal/mol; KI 2.57 nM), ZINC000043203371 (−12.2 Kcal/mol; KI 1.70 nM), ZINC000063933734 (−12.2 Kcal/mol; KI 296.76 pM), and ZINC000095092808 (−12.2 Kcal/mol; KI 4.02 nM) demonstrated the most favorable binding affinity coupled with an attractive inhibition constant. Moreover, these compounds exhibited consistent, strong, and stable interactions in MDS. These findings emphasize the need for further exploration of these compounds’ anti-tubercular properties, signaling their potential as leading candidates for TB treatment. They may spark additional research efforts focused on unraveling their mechanisms, ultimately establishing them as crucial molecules in the fight against TB.
Keywords: Drug resistance, Mycobacterium tuberculosis, Virtual screening, GlfT2, Drug targets, Inhibitors
Comparison of “Fascin” expression in various subtypes of lymphoproliferative disorders
Swati Sharma1 https://orcid.org/0000-0002-2808-8820 , Anuradha Kusum1, Mansi Kala1
1Department of Pathology, Himalayan Institute of Medical Science, Swami Rama Himalayan University, Dehradun, India.
E-mail: nareshchettri15@gmail.com
The introduction of the Fascin marker represents a significant advancement in lymphoma diagnosis, particularly in distinguishing Hodgkin’s lymphoma (HL) subtypes and non-Hodgkin’s lymphomas (NHL), including B-cell and T-cell variants. The study aimed to investigate Fascin expression in lymphoid tissues, both reactive and neoplastic, and to evaluate its variation across different lymphoma subtypes. This study conducted at Swami Rama Himalayan University’s Pathology department analyzed 63 lymphoma cases, revealing distinct Fascin expression patterns. Hodgkin’s lymphoma subtypes – Nodular Lymphocyte Predominant HL and Nodular Sclerosis classic HL (CHL) – showed 100% positivity, lymphocyte-rich CHL exhibited 50% positivity, and mixed cellularity CHL demonstrated 90% positivity. Among NHL cases, diffuse large B-cell lymphoma had 6% Fascin positivity, T-cell-rich B-cell NHL showed 50%, and anaplastic large cell lymphoma displayed the highest at 80%. Consistent Fascin positivity in specific lymphoma subtypes suggests its potential as a diagnostic marker, while its variable expression in other subtypes underscores the importance of considering molecular heterogeneity in lymphoma diagnosis for tailored therapeutic approaches. Further research into Fascin expression in lymphoma subtypes could enhance our understanding of disease biology and guide tailored therapeutic approaches.
Keywords: Fascin, Lymphoma, Hodgkin’s lymphoma, Non-Hodgkin’s lymphoma, Anaplastic large cell lymphoma
Isolation, molecular characterization, and preparation of exclusively lytic bacteriophages against Mycobacterium species
Garima Kandwal1, Tanmayee Nayak1, Anuja Kakkar1, Lav Kumar Jaiswal1 https://orcid.org/0000-0001-8287-3830 , Ankush Gupta1
1Department of Biochemistry, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India.
E-mail: nareshchettri15@gmail.com
Tuberculosis (TB), a chronic communicable disease affecting India and other low-middle-income countries, is caused by Mycobacterium tuberculosis. Antibiotics, administered over extended periods, serve as the fundamental way of TB treatment; however, the escalating antimicrobial resistance (AMR) necessitates the exploration of alternative therapies. Phage therapy, with its specificity, is deemed effective against stubborn bacterial infections. Mycobacteriophages, double-stranded DNA (dsDNA)-tailed viruses infecting Mycobacterium species, exhibit self-replication, high host specificity, and resilience to environmental factors, which are promising in combating AMR. Our study screens and isolates mycobacteriophages from diverse environmental samples, including the river Ganges. We have isolated mycobacteriophage from Banaras Hindu University (BHU) gate named Kashi BHU Gate 2 (KBG2), confirmed through spot assay. Genomic analysis of KBG2 unveiled a 42044 bp whole-genome sequence comprising 60 gene products, with 25 functionally annotated and 35 as hypothetical proteins, including an arginine-encoding tRNA. The identification of integrase and repressor genes indicates the temperate nature of the phage. Furthermore, restriction digestion with PstI, HindIII, and BamHI confirmed the phage dsDNA restriction profile. Structural determination using transmission electron microscopy reveals that KBG2 belongs to the Siphoviridae family. The genome map of KBG2 was generated using the CG view tool. Cluster analysis categorizes KBG2 into Cluster G1. Further, functional characterization demonstrates that KBG2 depicts stability across the 4°C–45°C temperature range and pH, i.e., acidic (pH 4) to alkaline (pH 9) range. The genetically engineered G cluster phage (BPs) has already demonstrated its clinical efficacy in treating Mycobacterium abscessus infections. Hence, G cluster phage holds immense potential for therapeutic applications, with the requisite modifications achievable through genetic engineering.
Keywords: Mycobacterium, Antibiotic resistance, Bacteriophages, Transmission electron microscopy, Siphoviridae family
Comparison of conventional and molecular methods for testing of Neisseria gonorrhoeae and determination of fluoroquinolone resistance in patients with sexually transmitted infections
Sangeeta Rawat1, Garima Mittal1, Rajender Singh1 https://orcid.org/0000-0002-2697-4765 , Geeta Rawat2, Sanjay Gupta3
1Department of Microbiology, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, 2Department of Microbiology, Dev Bhoomi Uttarakhand University, 3Himalayan School of Biosciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Gonorrhoea is the second most prevalent sexually transmitted infection (STI) worldwide caused by Neisseria gonorrhoeae. Due to the low sensitivity of the culture method, in recent times, molecular methods have been used more commonly to detect gonococci directly from clinical samples. Fluoroquinolone resistance in gonococci is due to alteration in the target site, in quinolone resistance determining regions of gyrA and parC. This study aims to compare the conventional and molecular methods to detect the prevalence of N. gonorrhoeae and the detection of fluoroquinolone resistance in clinical samples.
Materials and Methods: The present study was conducted from 2021 to 2023 in the Department of Microbiology. A total of 73 samples that were clinically suspected for STI were evaluated. The gonococci were directly detected from clinical samples using the polymerase chain reaction (PCR). The culture was done on GC agar with antibiotics and blood agar. The fluoroquinolone resistance was determined by the disc diffusion method and MIC determination was done by an E-test. Detection of fluoroquinolone resistance was done by detection of gyrA and parC genes by PCR.
Results: Out of 73 samples, 4 (5.5%) samples showed intracellular diplococci in Gram’s stain. Only 3(4.1%) samples showed growth of N. gonorrhoeae on culture media. These 3 isolates were resistant to ciprofloxacin. In molecular testing, 20 (27.4%) samples were positive for porA pseudogene, which confirms the presence of N. gonorrhoeae in clinical samples. Mutation in the gyrA gene and parC gene was seen in 100% and 95%, respectively.
Conclusions: To diagnose N. gonorrhoeae from clinical samples, molecular methods proved more useful than the culture-based method. The positive samples showed the presence of gyrA and parC gene, representing high ciprofloxacin resistance. For these reasons, there is now increased interest in developing systems for the non-culture-based detection of gonococci and its antimicrobial resistance mechanisms.
Keywords: Sexually transmitted infections, Neisseria gonorrhoeae, Quinolone resistance determining regions, Fluoroquinolone resistance
Serum miR-21 expression levels in pre and postoperative oral cancer and its association with clinical and histological parameters
Rajat kala1 https://orcid.org/0009-0000-2339-7459
1Cancer Research Institute, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Jolly Grant, Dehradun.
E-mail: nareshchettri15@gmail.com
Introduction: For research on oral carcinogenesis, the analysis of microRNAs expressed in oral squamous cell carcinoma (OSCC) provides a complex network of interest. miR-21 is overexpressed in a large number of solid tumors.
Objectives: The aim of our study was to find the impact of surgical excision on serum miR-21 expression level in OSCC cases and to establish a correlation with clinicopathological parameters.
Methodology: The study was an exploratory, prospective observational study. The study was conducted with proper institutional ethical approval at Cancer Research Institute, Himalayan Institute Medical Sciences, Jolly Grant, Dehradun. Fifty-six histologically confirmed OSCC cases along with healthy subjects as a control enrolled. The relative fold expression change was calculated by the Livak method (2−∆∆ct).
Results: 28 out of 56 samples showed high expression of miR-21 (P < 0.0001), while 27 cases showed downregulation in post-surgery cases (P < 0.004), and one sample showed the same level of expression. There were 18 patients in which the difference between pre and post-surgery samples was above 25 days; no significant change was found in those patients (P = 0.06), while 38 patients in which the difference was below 25 days also showed no significant value (P = 0.145). We found that the correlation between pre-surgery serum albumin and globulin ratio (A:G ratio) and perineural invasion with miR-21 is significant (P = 0.053). No other parameter showed any significant correlation with miR-21.
Conclusion: Our results indicate that in a few cases of OSCC, the miR-21 expression was reduced significantly (P < 0.004), while the OSCC association with tumor microenvironment variables such as mitotic figure, histological grading, presence of necrosis, and tumor dimension was not significant except with perineural invasion and A: G ratio. It can be concluded that perineural invasion and the A: G ratio have a direct influence on miR-21. It is important to note that the research on post-surgery miR-21 expression level in oral cancer is still evolving, and post-surgery miRNA expression is a promising area of research with the potential to improve the management of oral cancer.
Keywords: microRNA, Perineural invasion, Clinic-pathological parameters, Tumor necrosis, etc.
Isoniazid mono-resistant tuberculosis: A challenge for tuberculosis eradication program
Vikram Sethi1 https://orcid.org/0000-0002-2673-3080 , Aarti Kotwal1, Varuna Jethani2
Departments of 1Microbiology and 2Respiratory Medicine, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Background: Antimicrobial resistance (AMR) is a major contributor to morbidity and mortality worldwide. The burden of drug-resistant tuberculosis (TB) is highest in India and one of the major concerns currently. With rifampicin (RIF) resistance considered as a surrogate marker for multidrug-resistant-TB, all efforts are focused on RIF resistance, leading to gradual escalation of isoniazid (INH) resistance.
Aim: A 10-year retrospective analysis of the burden of INH mono-resistance at our tertiary care center with the algorithm of TB resistance screening in place was done.
Methodology: We analyzed the trends of INH resistance pattern from June 2012 to October 2022 at our tertiary care hospital in north India. The clinical samples, both pulmonary and extrapulmonary, were genotypically evaluated using polymerase chain reaction followed by reverse hybridization technique (line probe assay [LPA]), and results with valid interpretation were enrolled in the study.
Results: Around 772 samples, pulmonary (58.8%) and extrapulmonary (41.2%), were processed during the 10-year time frame for LPA. We found that a total of 36% of samples were positive for Mycobacterium tuberculosis. Among the positives, 30.8% were resistant to RIF/INH or both. Among resistant cases, 21% were old, while 79% were newly diagnosed. We found 13.8% resistance to both RIF and INH in our study. The percentage of resistance was almost equal in both pulmonary and extrapulmonary. The total percentage of INH resistance among the TB cases was 8.7% in 10 years, but what was noticeable was a gradual decline in INH mono-resistance from 28.5% to 5.9% from 2012 to 2022.
Conclusion: An association between INH resistance screening and decreasing trend in INH mono-resistance has been witnessed in our pilot study. Detection of INH mono-resistant TB is essential for TB eradication programs.
Keywords: Isoniazid mono-resistant tuberculosis, Genotypic analysis, Line probe assay, Tuberculosis eradication, Tuberculosis-resistant screen
Home-made pickles: A source of community-acquired antimicrobial resistance
Aafreen1, Anjali Jaiswal1, Arthana Nair1, Bhavna Parmar1, Ram Karan1, Ashwini Chauhan1
1Department of Microbiology, University of Delhi, New Delhi, India.
E-mail: nareshchettri15@gmail.com
Pickling is an ancient Indian tradition practiced for thousands of years and continues today. We investigate the microbiological complexity of typical Indian homemade lemon pickles and non-vegetarian pickles made from dried prawns and fish aged 1–1.5 years as a potential source of antimicrobial resistance. Slurries with different salt concentrations, ranging from 0.9% to 15% (w/v) sodium chloride, revealed varied microbial communities. The isolates were analyzed based on morphology, biochemical characteristics, and enzyme activity. The isolates demonstrated a range of activities, including amylase, cellulase, xylanase, lipase, urease, proteinase, oxidase, catalase, and nitrate reductase. Molecular approaches, such as 16S ribosomal ribonucleic acid sequencing, provided more profound insights into microbial diversity. In addition, the biofilm-forming ability of these isolates was analyzed. For lemon pickle isolates, antibiotic sensitivity testing revealed ampicillin resistance in 0.9% and 9%, as well as streptomycin resistance in 6% and 9% of saline cultures. However, no resistance was observed for tetracycline hydrochloride and chloramphenicol. Isolates from fish and prawn pickles, grown at 0.9% salinity, showed resistance to ampicillin, streptomycin, and tetracycline hydrochloride. At 3% salinity, both pickle samples were resistant to ampicillin and streptomycin, with the prawn additionally showing resistance to tetracycline and chloramphenicol. Higher salinity levels (6–15%, w/v NaCl) were associated with increased resistance, notably to ampicillin and streptomycin, in fish and prawn samples. This study highlights the potential significance of pickled foods in transmitting antibiotic-resistant genes. It emphasizes the necessity of understanding microbial dynamics in traditional food preservation methods and advocates for responsible antibiotic use in food production to prevent community-acquired antimicrobial resistance.
Keywords: Home-made pickles, Antibiotic resistance, Microbial diversity, Biofilm formation
Environmental BL tester: A method and protocol to detect antimicrobial resistance in environmental samples
Tanmoy Sen1 https://orcid.org/0009-0007-7442-6441 , Saugata Hazra1,2
1Department of Bioscience and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Haridwar, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) poses a great threat to humanity, like a curse of our advancement in science to fight against microbial infection across the globe. Bacterial resistance is spreading like wildfire not only in medical settings but also in the environmental realm by the expression of different genes to bypass the effect of antimicrobials; beta-lactamase appears to be the major contributor among those. AMR impacts human health and jeopardizes the fundamental pillars of a country’s or continent’s economic status by causing harm in four domains: Human, animal, food, and environmental. Environmental BL-Tester is a method to detect the resistant bacterial load, having beta-lactamase, within 25–30 min from environmental samples, including wastewater, soil, and river/lake water, which serves as significant sources for the escalating emergence of drug-resistant bacteria due to improper discharge. Drainage and sewage water samples were collected from Roorkee district, Haridwar, India. Growth of the drug-resistant bacteria in respective samples was estimated by growth inhibition assay where ampicillin (10 μg/mL’ CLSI 2022) was used to select the resistant bacteria. A dye named nitrocefin was used to detect the presence of enzyme-mediated drug resistance due to its property of chromogenic shift (390–486 nm) visible to the naked eye. It works with four filters that work, respectively, by sieving out particles from large to small and finally adhering to bacterial cells. The overall assay is very easy to perform, can generate results within a narrow detection time, and is very specific toward its target to avoid false-positive outcomes.
Keywords: AMR, beta-lactamase, nitrocefin, environment
Biofilms: Understanding molecular mechanism, genetics, and innovative control strategies
Kumar Sachin1 https://orcid.org/0000-0002-9719-5611 , Tanisha Bansal1, Santosh Kumar Karn2
1Himalayan School of Biosciences, Swami Rama Himalayan University,
1Department of Biochemistry and Biotechnology, Sardar Bhagwan Singh University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
A significant counter to the successful operation of maritime industrial units, medical implant devices, or any wastewater treatment plant is the formation of biofilm on the surfaces of tanks and tubes. The observed phenomenon of biofilm development is ubiquitous as part of microbial growth on any surface. The robust and resistant nature of biofilms confers great advantage to the microbial community with regard to any antimicrobial strategy targeted against them, hence contributing to their antimicrobial resistance. Although biofilms are reported to have been positively applied in the development of biodiesel, biofertilizer, and water pollutant degradation, they are overall a menace to human society and cause considerable economic loss to industrial units. Numerous approaches to disrupt the formation of biofilms have been proposed; however, it remains a grave area of concern with little results. The focus of this article would be to understand the latest insights into the molecular mechanism of biofilm formation, its genetics and metabolic activity within the structure, and innovative ways in which the problem can be resolved. To develop effective anti-biofilm strategies, it is imperative to understand how biofilms are self-synthesized by microorganisms, the active genes in the process, and biochemical pathways involved in the formation of such complex organized 3D architecture, which shields the microbial cells from any kind of environmental stress factors.
Keywords: Biofilm, Molecular mechanism, Genetics, Metabolism, Control
Pattern of antimicrobial sensitivity of clinical isolates of Pseudomonas aeruginosa in a tertiary care center
Muskan Khurana1, Barnali Kakati1 https://orcid.org/0000-0002-1592-9109 , Nupur Koul, Neha1
1Department of Microbiology, Himalayan School of Biosciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Pseudomonas aeruginosa stands as a prominent culprit behind nosocomial infections globally with a concerning surge in carbapenem-resistant P. aeruginosa (CRPA) posing significant therapeutic challenges.[2] Detecting carbapenemase-producing organisms holds crucial importance in guiding therapy and curtailing dissemination. Phenotypic tests such as the modified carbapenem-inactivation method (mCIM) and ethylenediaminetetraacetic acid-modified carbapenem-inactivation method (eCIM) have emerged as valuable tools for identifying and distinguishing between serine and metallo-based carbapenemase.
Aims and Objectives: The aims and objectives of the study are to identify CPRA among various clinical isolates, determine their antimicrobial sensitivity profile, detect Carbapenemase producers by phenotypic methods (mCIM, eCIM),[4] and check the sensitivity of these isolates against ceftazidime-avibactam using ETest method.
Materials and Methods: This observational study was conducted in Department of Microbiology, Himalayan Institute Medical Sciences over a period of 2 months. A total of 87 consecutive, non-duplicate isolates of P. aeruginosa were included and subjected to antimicrobial sensitivity testing and carbapenemase detection using mCIM and eCIM methods.
Results: Out of 87, 29 (33%) were identified as CRPA. CRPA isolates were found resistant to other antibiotics such as cefoperazone-sulbactam (86%), ciprofloxacin (86%), aztreonam (66%), and gentamicin (66%). Out of 29 CRPA, 5 (17%) were identified as carbapenemase and 24 (83%) as carbapenemase non-producers. Out of 5 carbapenemase producers, 04 (80%) were identified as metallo β-lactamase and 01 (20%) serine carbapenemase producer. All metallo-beta-lactamases-producing CRPA were found sensitive to ceftazidime-avibactam.
Conclusion: Findings of this study suggest high resistance rates among CRPA isolates for more than one antimicrobial class. CRPA lacked detectable carbapenemase activity which indicates other drug resistance mechanisms as reasons for carbapenem resistance.
Keywords: Pseudomonas, Resistant, Carbapenemase, Antimicrobial, Ceftazidime-avibactam
Antimicrobial susceptibility pattern of eight drugs against Helicobacter pylori strains isolated from patients with gastrointestinal diseases in North India
Safiya Arfi1, Prateek Sharma1, Mithun Kumar2, Ashwini Setya2, Shubham Mehra1, Kunal Das3, Rajashree Das1 https://orcid.org/0000-0003-1202-4942
1Centre for Medical Biotechnology, Amity Institute of Biotechnology, Amity University, Noida, Uttar Pradesh, 2Department of Gastroenterology and Hepatology, Max Super Speciality Hospital, New Delhi, 3Department of Gastroenterology, Yashoda Super Specialty Hospital, Ghaziabad, Uttar Pradesh, India.
E-mail: nareshchettri15@gmail.com
Objectives: This analysis aimed to assess the antimicrobial susceptibility of eight drugs against Helicobacter pylori strains and investigate the genetic diversity of H. pylori virulence markers to predict clinical outcomes in North India.
Materials and Methods: Tissue biopsy samples from 180 patients with gastrointestinal diseases were examined for H. pylori presence using microbial culture, rapid urease test, and polymerase chain reaction. Minimum inhibitory concentrations (MICs) of antibiotics were determined through the agar dilution method. Statistical analysis utilized R STUDIO and Statistical Package for the Social Sciences 20.0 software.
Results: Among 180 samples, 58 H. pylori strains were cultured. Drug resistance prevalence was observed against cefixime (CFM) (41.3%), furazolidone (FZD) (34.4%), amoxicillin (AMX) (20.7%), levofloxacin (LVFX) (70.7%), metronidazole (MTZ) (39.6%), tetracycline (TET) (20.7%), clarithromycin (CLA) (17.2%), and rifabutin (RIF) (17.2%). Genotypic variations were noted in H. pylori strains. Various resistance patterns were identified, including single-drug resistance (21%), dual resistance (29.3%), triple resistance (18.9%), and multidrug resistance (25.8%). Resistance rates in MTZ, CLA, and RIF were significantly higher in females compared to males (P = 0.005, P = 0.002, and P = 0.02, respectively). TET and LVFX resistance levels were significantly higher in gastritis compared to other disease groups (P = 0.04 and P = 0.0003, respectively).
Conclusion: This study is the first in North India to report H. pylori antimicrobial resistance against RIF and CFM. TET, AMX, CLA, and RIF were found to be more effective antibiotics against H. pylori infections, with an observed increase in LVFX resistance in North India.
Keywords: Helicobacter pylori, Antibiotic resistance, Levofloxacin, Clarithromycin, Rifabutin, Virulence genes
Assessing the influence of the virulent gene of Helicobacter pylori on gastric microbiota dynamics
Aditya Vikram Singh1 https://orcid.org/0000-0003-1202-4942
1Centre for Medical Biotechnology, Amity Institute of Biotechnology, Amity University, Noida, Uttar Pradesh, 2Department of Gastroenterology, Yashoda Super Specialty Hospital, Ghaziabad, Uttar Pradesh, 3Department of Psychiatry, Indian Council of Medical Research, 4Department of Psychiatry, All India Institute of Medical Sciences, New Delhi, India.
E-mail: nareshchettri15@gmail.com
Background: Helicobacter pylori has several virulence factors, such as cytotoxin-associated gene A (cagA) and the, induced by contact with epithelium antigen (iceA). H. pylori infection causes several chronic gastrointestinal diseases. Our study elucidates the relationships between the cagA status of H. pylori-infected patients and the compositional changes in the gastric microenvironment of patients suffering from gastrointestinal diseases.
Methods: 35 patients who attended the Department of Gastroenterology, Max Super Specialty Hospital, Vaishali, Ghaziabad, Uttar Pradesh, were included in this study. Their gastric biopsies were collected, and cag A genes of H. pylori were amplified using polymerase chain reaction.
Results: A significant difference in the alpha and beta diversity indexes was found between the study groups. Helicobacter, Staphylococcus, Rothia, Dialister, and Faecalibacterium showed a significant correlation in the disease group as compared to the Hp −ve patients. The microbial biomarkers were identified; 29 were found to be statistically significant (P < 0.05) in the Hp +ve/Cag A +ve group, whereas 19 were found to be statistically significant (p-value < 0.05) in the Hp +ve/Cag A −ve group. The co-occurrence network analysis showed a significant association between the Gram-negative bacterial genera in both Hp +ve groups; however, the significance level increased due to the presence of the cagA gene.
Conclusion: This research unravels the intricate relationship between, the presence of the cagA gene during H. pylori infection, and the gastric microbiota. Distinct microbial compositions were observed at both phylum and genus levels, delineating the impact of H. pylori infection and cagA genotypic variations. However, the clinical relevance and the mechanism underlying the altered microbiota composition require further functional studies.
Keywords: Helicobacter pylori, cagA gene, Gastric microbiota, Dysbiosis
To study the disease profile and outcomes of multidrug-resistant patients in the surgical ward at a tertiary care hospital
Manish Kulshrestha1 https://orcid.org/0000-0002-3207-072X , Deepak Rajput1
1Department of Microbiology, All India Institute of Medical Sciences (AIIMS), Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Multidrug-resistant organisms (MDROs) are microorganisms, mainly bacteria that are resistant to two or more classes of antimicrobial agents. There are very few longitudinal observational studies on multidrug-resistant (MDR) infection among surgical patients. In this longitudinal observational study, we aim to study the disease profile and outcomes of MDR patients in the surgical ward.
Objectives: (i) To estimate the incidence of MDR infection in the surgical ward. (ii) To assess the demographical, clinical, laboratory, and treatment profiles of MDR patients in the surgical ward. (iii) To assess the outcomes of MDR patients in the surgical ward.
Methods: The data entry operator notified MDR reporting through a live Excel sheet, which was followed by a project investigator in the surgery ward. All relevant data were collected using a scheduled list, and the patient was followed during the hospital stay.
Results: Seven (5.14%) patients died out of 136 MDR patients under evaluation from August 2023 to February 2024 and 123 (90.44%) were discharged after recovery while six (4.41%) were discharged on request. The MDR organisms isolated were Klebsiella (36.02%), Escherichia coli (33.08%), Acinetobacter (10.29%), Enterococcus (6.61%), Pseudomonas (2.94%), and others (8.82%), out of 136 samples. E. coli was the most common MDRO in body fluid samples while Klebsiella was the most common MDRO in urine and blood samples. Antimicrobial use as per the World Health Organization AWaRe classification, Access (67.14%), Watch (16.9%), Reserve (15.94%).
Conclusion: The most common MDRO in surgical ward was Klebsiella pneumonia followed by E. coli. Mortality of MDR patients was 5.14% in surgical ward.
Keywords: Multi-drug resistance, infection, antibiotic stewardship
An insight toward the extravagant change in the positioning of a tryptophan residue present in the active site of a high-resolution crystal structure of a mutant of metallo-β-lactamase IMP-1: A carbapenemase rapidly disseminating globally
Subhecchha Baidya1 https://orcid.org/0009-0005-4852-246X , Saugata Hazra1,2
1Department of Bioscience and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Haridwar, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) has gradually widened enough to negate the advances in medical sciences successfully. IMP-1, a bacterial enzyme, belongs to metallo-beta-lactamases (MBL), a subclass of beta-lactamase (BL), inactivates beta-lactam drugs by hydrolyzing the B-lactam bond (amide bond), the most prescribed antibacterial. IMP-1 and its mutant have been cloned, over-expressed, purified to a single band in SDS-PAGE, and subjected to biophysical-biochemical characterization as well as in vivo (MIC and disc diffusion assay) experiments with the determination of crystal structure by XRD. Steady-state kinetic (SSK) experiments suggest IMP-1 and its mutant has a specific affinity toward different drugs even in the same group (e.g., Penicillin). Circular dichroism (CD) experiments confirmed its proper folding after which SSK profile showed it cleaves the first-generation cephalothin more readily with a kcat/Km value of 30.52 μm/s and surprisingly shows higher kcat values for doripenem compared to imipenem and ertapenem. Inhibition profiles are thoroughly examined by Isothermal Titration Calorimetry (ITC) as MBLs currently have only Aztreonam as an inhibitor. Inhibitors (e.g.- L-Captopril, Dipicolinic acid, and Cisplatin) are observed to show promising results which could be a partial success of this study. XRD experiments were carried out for the mutant and were solved at 2.9Å using IMP-1 as a template with an Rfree of around 0.27 after refinement. Interestingly, the mutant’s structure revealed an essential Tryptophan residue is covering the active site as a lid, unlike IMP-1.
Keywords: Antimicrobial resistance, metallo-beta-lactamases, IMP-1, XRD, steady state kinetics, circular dichroism, isothermal titration calorimetry.
A system, method, and device for the detection of drug-resistant pathogen: Advanced BL tester
Harshita Sharma1 https://orcid.org/0009-0005-6685-9664 , Kunal Dhankhar1, Subhecchha Baidya1, Rajsekhar Adhikary1, Saugata Hazra1,2
1Department of Bioscience and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Beta-lactams stand as the most indispensable antibiotics throughout human history. However, their efficacy has been challenged by the expression of beta-lactamases in various organisms. These enzymes fall into two main categories: Serine-based and metallo beta-lactamases, capable of hydrolyzing all beta-lactams, including carbapenems, often reserved as antibiotics of last resort. Pathogens such as Klebsiella pneumoniae and Pseudomonas aeruginosa express diverse beta-lactamases, leading to resistance against beta-lactams. Hence, rapid beta-lactamase detection in clinical samples is paramount for tailoring patient treatment regimens. This study presents a novel method and device for the simultaneous detection of Serine and Metallo beta-lactamases. The device comprises three key units. The first unit, the Shaking, Heating (SHT) unit, facilitates the growth of clinical samples for detection. The second unit, the BL tester body, houses a bacterial filter that concentrates bacteria from the media. The third unit is the display cassette, enabling visualization of results. Notably, this device can differentiate between the presence of Serine and Metallo beta-lactamases. It employs Avibactam (a diazabicyclooctane) and ethylenediaminetetraacetic acid (EDTA) (Chelator) for differential detection. Avibactam inhibits Serine beta-lactamases through Carbamylation and Decarbamylation, while EDTA inhibits Metallo beta-lactamases through the chelation of Zinc. In addition, the system can detect the presence of an Efflux pump. With this innovative system, beta-lactamase detection can be accomplished within 2–3 h, greatly expediting the determination of appropriate antibiotic regimens for patients.
Keywords: Beta lactamase, detection, antimicrobial resistance
Multidrug-resistant patients in the surgical ward at a tertiary care hospital – A longitudinal study
Manish Kulshrestha1 https://orcid.org/0000-0002-3207-072X , Prasan Kumar Panda2, Balram Ji Omar3, Deepak Rajput1
Departments of 1Surgery, 2Internal Medicine, 3Microbiology, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Multidrug-resistant organisms (MDROs) are microorganisms, mainly bacteria, that are resistant to two or more classes of antimicrobial agents. There are very few longitudinal observational studies on MDR infection among surgical patients. In this longitudinal observational study, we aim to study the disease profile and outcomes of MDR patients in the surgical ward.
Objectives:
To estimate the incidence of MDR infection in the surgical ward
To assess the demographical, clinical, laboratory, and treatment profiles of MDR patients in surgical ward.
To assess the outcomes of MDR patients in surgical ward
Methods: The data entry operator notified MDR reporting through a live excel sheet, which was followed by a project investigator in the surgery ward. All relevant data was collected using a scheduled list and patient was followed during the hospital stay.
Results: Seven (5.14%) patients died out of 136 MDR patients under evaluation from August 2023-February 2024 and 123 (90.44%) were discharged after recovery while six (4.41%) were discharged on request. The MDR organisms isolated were Klebsiella (36.02%), E.coli (33.08%), Acinetobacter (10.29%) , Enterococcus (6.61%), Pseudomonas (2.94%) and others ( 8.82%), out of 136 samples. E.coli was the most common MDRO in body fluid samples while Klebsiella was the most common MDRO in urine and blood samples. Antimicrobial use as per WHO AWaRe classification, Access (67.14%), Watch (16.9%), Reserve (15.94%).
Conclusion: Most common MDR organisms in surgical ward were Klebsiella pneumonia followed by E. coli. Mortality of MDR patients was 5.14% in surgical ward.
Keywords: Multidrug resistant infections, antibiotic stewardship, surgical infections
A matched case–control study to determine the risk factors for carbapenem-resistant Klebsiella pneumoniae infection and its outcome in a tertiary care hospital
Manish Kulshrestha1 https://orcid.org/0000-0002-3207-072X , Anita Nandi2
1Department of Surgery, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India 2Department of Microbiology, Medical College and Hospital, Kolkata, West Bengal, India.
E-mail: nareshchettri15@gmail.com
Introduction: Carbapenems are frequently used to treat infections due to extended-spectrum β-lactamase-producing Klebsiella pneumoniae. Thus, the emergence of infections due to carbapenem-resistant K. pneumoniae (CRKp) is a major public health concern. This study aimed to evaluate the risk factors of CRKp infections so that appropriate intervention strategies can be developed. The importance of resistance development was highlighted by exploring the outcome of such infections in terms of duration, cost, and effectiveness of treatment.
Methodology: We conducted two matched case-control studies comparing CRKp cases with controls and CSKp cases with controls. The controls were selected among patients showing no growth on culture. Data were collected from all available medical records, proper history taking, and follow-up with the help of pre-designed and pre-tested schedules.
Results: Recent (≤7 days) invasive procedures such as urinary catheterization (odds ratio [OR] = 11.56, 95% confidence interval [CI]: 2.373854–60.4589, P = 0.0003), parenteral nutrition (OR = 7.88, 95% CI: 0.7937471–379.0258, P = 0.0393), pulmonary comorbidities (OR = 7.28, 95% CI 1.617428–35.02413, P = 0.0025), CNS disease (OR = 14.53, 95% CI 1.590895–666.6596, P = 0.0040), diabetes mellitus (OR = 10, 95% CI 1.637118–103.0252, P = 0.0029), and recent (≤1 month) special treatments (OR = 7.88, 95% CI 0.7937471– 379.0258, P = 0.0393) were identified as risk factors for carbapenem-resistant K. pneumoniae infection. Previous use of carbapenem showed the highest association (OR = 14.53, 95% CI 1.590895–666.6596, P = 0.004) with CRKp infections. Mortality, hospital stay, and cost of treatment were higher in CRKp cases than in CSKp cases. Antibiotic regimens containing carbapenem and piperacillin/tazobactam had the highest mortality (13.63%).
Conclusion: Patients exposed to the risk factors for CRKp infections should be timely screened for signs of infection and promptly treated with appropriate antibiotic regimens. Carbapenem should not be included in empirical treatments for infections to decrease the emergence of CRKp. This would decrease the mortality; hospital stay, and cost of treatment of the patients.
Keywords: Multidrug-resistant infections, Antibiotic stewardship, Multidrug resistant infections, antibiotic stewardship, carbapenem resistant Klebsiella pneumoniae
Trends in the antimicrobial resistance among bacterial uropathogens at Dr. Susheela Tiwari hospital, Haldwani
Sukriti Chandra1 https://orcid.org/0009-0003-4732-4221 , Umesh1, Shambhavi Singh1, Neha Kaushal1, Mayuri Srivastava1
1Department of Microbiology, Government Medical College, Haldwani, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Background: Urinary tract infections (UTIs) are one of the leading causes of morbidity, mortality, and health-care expenditure. Antimicrobial resistance (AMR) is a worldwide health concern that results in limited treatment options and poor outcomes. This study was conducted at Dr. Susheela Tiwari Hospital, Haldwani, to identify the bacteriological profile and antibiotic resistance patterns among patients with UTIs.
Material and Methods: The study was retrospective and analyzed 8440 samples of urine culture received at the Department of Microbiology, Government Medical College Haldwani from January 1, 2022, to December 31, 2023. Data were analyzed based on age, gender, identification of culture isolates, and their antibiotic resistance pattern.
Result: Of the total 8440 urine samples received, 1115 were positive for pathogenic bacteria (13.2%) with female preponderance (59%). The highest incidence of UTI cases was in the age group 0–15 years (36.8%). The most common uropathogen was Escherichia coli (62.5%), followed by Enterococcus spp. (15.6%) and Klebsiella spp. (10.5%). In Gram-negative isolates, penicillin, cephalosporins, and fluoroquinolones showed high resistance, while carbapenems and fosfomycin showed the least resistance. Most Gram-positive isolates were resistant to ciprofloxacin and ampicillin whereas gentamicin and cotrimoxazole were the least resistance.
Conclusion: E. coli was the most prevalent Gram-negative uropathogen and carbapenems, fosfomycin, and colistin were effective antibiotics. Among Gram-positive uropathogen, Enterococcus spp. was most frequent isolate and vancomycin and linezolid were most effective. Determining local antibiotic resistance patterns of uropathogens helps in rational empirical therapy and sustains antimicrobial stewardship programs.
Keywords: Urinary tract infections, Antimicrobial resistance, Uropathogens, Empirical therapy, Antimicrobial stewardship
Understanding antimicrobial resistance: Insights into bacterial mechanisms and strategies for intervention
Renu Chaudhary1 https://orcid.org/0000-0001-7261-7091 , Vivek Kumar1
1Himalayan School of Biosciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) has emerged as a burgeoning global crisis, jeopardizing the efficacy of antimicrobial therapies across medical care, animal treatment, and disease reduction. This escalating challenge underscores the imperative for transformative action. While the natural development of antibiotic resistance in microbes is a typical occurrence, the accelerated pace of this process can be attributed to heightened exposure to antibiotics in the environment, health care, and agriculture. Emphasizing the need for intervention, we delve into the genetic underpinnings, unraveling the roles of specific genes and mutations in bacterial populations that drive resistance. Resistance mechanisms, including efflux pumps, biofilm formation, and enzymatic degradation, provide a nuanced view of bacterial strategies against antimicrobial agents. Genomic insights spotlight the evolutionary pathways of resistance genes, showcasing the diversity of mechanisms across bacterial species through comparative genomics. Horizontal gene transfer emerges as a critical facilitator in the spread of resistance, examining the role of mobile genetic elements such as plasmids and transposons. Strategies for combating AMR are outlined, with a focus on antimicrobial stewardship programs and the imperative role of global surveillance systems in tracking and monitoring trends in AMR. The current advancements feature innovative approaches such as phage therapy, CRISPR-based technologies, and the promise of nanotechnology in developing novel antimicrobial agents. These interventions offer hope in overcoming the challenges posed by AMR. Concluding with an analysis of current challenges, including diagnostic limitations and policy implementation hurdles, the review outlines future prospects and emerging research areas.
Keywords: Antimicrobial resistance, multidrug resistance, Resistance mechanisms, Horizontal gene transfer, Nanotechnology, Global health
Advanced technological methods for characterizing Mycobacterium tuberculosis
Mayank Purohit1, Brijesh Kumar1, Anjali1, Sheetal1, Neha Chauhan2 https://orcid.org/0000-0002-1687-0831
Departments of 1Medical Laboratory Technology and 2Microbiology, School of Paramedical and Allied Health Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The tubercle Bacillus is difficult to combat because it can remain latent in the human body for many years, grow slowly when compared to other bacteria, and cause subtle and gradual symptoms at first. Recent advances in mycobacterial genomics and human cellular immunology have resulted in two new blood tests that detect tuberculosis (TB) infection by measuring in vitro T-cell interferon (IFN) gamma release in response to two unique antigens that are highly specific for Mycobacterium tuberculosis. Interferon-gamma release assays (IGRAs) are blood tests that can help diagnose TB infection. IGRAs measure the amount of interferon-γ produced by lymphocytes after stimulation with two or three antigens that are specific to M. tuberculosis. These antigens are not found in the Bacillus calmette-guerin vaccine or in environmental mycobacteria. IGRAs can be used in place of tuberculin skin tests in certain situations, such as contact investigations, testing during pregnancy, and screening of health-care workers. Rapid and accurate diagnosis of symptomatic individuals is a key component of global TB control strategy. Remarkable progress has lately been made in affluent countries to improve the speed and quality of mycobacteriology diagnostic services, but for the majority of the world, where TB remains a major public health concern, these benefits have yet to be realized. New TB diagnostics are urgently needed to replace or enable acid–fast bacilli (AFB) microscopy for identifying smear-positive patients, as well as to improve AFB smear-negative case detection for usage in low-income countries.
Keywords: Tuberculin skin test, Interferon, Acid–fast bacilli, Bacillus calmette-guerin, Interferon-gamma release assays, Mycobacteriology
Development of herbal and antibiotic-loaded hydrogel dressing against antimicrobial resistance pathogen in wound healing
Komal Singh1, Narayan Chandra Mishra2, Saugata Hazra2,3 https://orcid.org/0000-0002-3074-1534
Departments of 1Polymer and Process Engineering and 2Bioscience and Bioengineering and Polymer and Process Engineering, Indian Institute of Technology, 3Center for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
In recent years, wound healing process has slowed due to the increased burden of pathogens resistant to antimicrobials. Wound infection with antimicrobial resistance (AMR) leads to a higher mortality and morbidity leading to an increase in health-care costs and affects the wound management system significantly. Owing to the increasing AMR, the appropriate use of antibiotics is vital to prevent and manage wound infection. We are developing an antibiotic cocktail and herbal extract-loaded wound dressing material to combat AMR-conjugated wound infection. This wound dressing is composed of natural as well as synthetic polymers such as collagen and polyvinyl alcohol along with plant extracts and cocktail drugs. The morphological, rheological, functional group, and in vitro degradation studies were done by field emission scanning electron microscopy, modular compact rheometer, and Fourier transform infrared spectroscopy. The water retention, swelling, and percentage of drug release at the fixed time opted as the dependent variable, interaction with NIH3T3 mouse skin fibroblasts, their antimicrobial efficacy against Gram-positive Staphylococcus aureus (S. aureus) and Gram-negative Escherichia coli bacteria, methicillin-resistant S. aureus, Klebsiella pneumoniae carbapenemase, Pseudomonas aeruginosa, Acinetobacter baumannii. The present study concludes that the dressings are cytocompatible and effective against all strains of bacteria tested to date, with the cocktail antibiotics showing higher efficacy. These wound dressings should show up as a promising alternative to treat AMR-infected wounds.
Keywords: Antimicrobial, wound, infection
Understanding the chemistry of carbapenem hydrolysis by SME-1 class A carbapenemase from Serratia marcescens
Kunal Dhankhar1,2, Saugata Hazra1,3 https://orcid.org/0000-0002-3074-1534
1Department of Bioscience and Bioengineering, 2Department of Polymer and Process Engineering, Indian Institute of Technology, 3Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
In the past few years, there have been several outbreaks reported on the Serratia marcescens, particularly in the Neonatal Intensive Care Units, which cause septicemia and other deadly infections. β-lactams remain the major antibiotics used to treat this infection. However, the organism has acquired resistance to β-lactams by expression of an Ambler Class A Carbapenemase SME-1. This enzyme can cleave virtually all of the beta-lactams available today, rendering them inactive. In this study, we have done the crystallization of SME-1 followed by soaking with diazabicyclooctanes such as avibactam and relebactam to understand the mechanism of binding of these inhibitors. Further to understand the mechanism of carbapenem hydrolysis, SME E166A mutant was designed and purified. It was followed by crystallization and soaking with carbapenems and higher-generation cephalosporins. High-resolution crystal structures were obtained with biapenem, cefpirome, ceftaroline, and other carbapenems. SME E166A Acyl Enzyme complex with biapenem showed the presence of imineenamine tautomers in the crystal structure that was confirmed in solution by NMR spectroscopy. From enzyme kinetics, it was found that MBIs are inhibitors of SME-1. Isothermal titration calorimetry of SME E166A with carbapenems showed that meropenem, ertapenem, and biapenem were poorer binders (KD in the range of 10−7 M) than the rest carbapenems (KD in the range of 10−8M). Enzyme kinetics also showed that SME-1 has a lower catalytic efficiency (kcat/ Km~1) toward biapenem. This information clearly demonstrates the role of Imine-Enamine tautomerism in carbapenem hydrolysis and the chemical groups required to inhibit the enzyme that will be used in drug discovery.
Keywords: Antibiotics, septicaemia, carbapenemase
Necrotizing bacterial pneumonia – A mimicker of relapse of pulmonary tuberculosis
B. Darshan1, Ritu Sangwan1, Prasan Kumar Panda2 https://orcid.org/0000-0002-3008-7245
1Department of Internal Medicine (ID Division), All India Institute of Medical Sciences, 2Department of Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Early and accurate diagnosis plays a crucial role in preventing antimicrobial resistance (AMR) by ensuring appropriate treatment. This case study highlights the importance of distinguishing between necrotizing bacterial pneumonia and tuberculosis (TB) relapse, as it significantly impacted the management and prevented potential tubercular AMR. The patient is a 41-year-old woman, a healthcare worker, with a history of pleural TB, 2-year back for which she had completed 9 months of anti-tubercular therapy (ATT), presented with complaints of abdominal pain in her right upper and lower quadrants for 2 months, swelling of both lower limbs with abdominal distension for 1 month, and worsening shortness of breath for 20 days. The patient was admitted outside hospital and was started on ATT suspecting the TB relapse. There was no improvement after 15-day ATT and was referred to higher center. In emergency, with the possibility of disseminated TB, ascitic tapping was done and patient was admitted in medicine ward. Her ascitic fluid analysis showed features of cardiac ascites with signs of congestive hepatopathy. ATT was stopped. Her procalcitonin levels were raised and contrast-enhanced computed tomography (CECT) thorax revealed mediastinal necrotizing lymphadenopathy encasing the right main pulmonary artery and causing severe luminal attenuation explaining the anasarca due to right-sided heart failure and showed necrotizing pneumonia and right-sided empyema. Patient was started on empirical antibiotics along with heart failure treatment and pleural fluid analysis showed exudative picture with negative CBNAAT and normal ADA. BAL fluid and EBUS TBNA’s CBNAAT were negative and bacterial cultures came positive for Serratia marcescens. Patient responded well to the culture-guided antibiotic therapy with significant clinical improvement and repeat CECT thorax, 15 days follow-up, showed decrease in lymph node size and number, assuring the diagnosis of necrotizing bacterial pneumonia in a case of old TB. In conclusion, accurate diagnosis of necrotizing bacterial pneumonia over TB is paramount in preventing AMR. This case signifies the significance of right diagnosis and right treatment in optimizing patient management, reducing AMR risks, and improving clinical outcomes.
Keywords: Antimicrobial resistance, Cardiac ascites, Necrotizing lymphadenopathy, Pneumonia, Serratia marcescens
Delving into the depths: A multifaceted in silico investigation of natural variants of class A beta-lactamase cefotaximase - munich, illuminating the molecular mechanisms underlying the emergence of novel antibiotic resistance
Deepika Singh1, Shreya Sharma1, Devesh Bhimsaria2, Arup Samanta1,3, Saugata Hazra2,3 https://orcid.org/0000-0002-3074-1534
Departments of 1Physics and 2Biosciences and Bioengineering, Indian Institute of Technology, 3Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antibiotics have undoubtedly been a game-changer in health care. However, their widespread use has led to a vast army of beta-lactamases in microorganisms, sparking the notorious issue of antimicrobial resistance. These sneaky beta-lactamases chop up the amide bond in beta-lactam-based antibiotics, rendering them as effective as a chocolate teapot and starring in causing those pesky nosocomial infections. In the early 1980s, third-generation cephalosporins emerged, ready to take on beta-lactamase-mediated bacterial diseases. However, alas, their overuse paved the way for the appearance of extended-spectrum beta-lactamases (ESBLs), particularly fond of hanging out in Enterobacteriaceae, the party animals responsible for urinary tract infections and bloodstream infections. Then, in the late 1980s, a plasmid-mediated class-A ESBL strutted onto the stage with a talent for cleaving cefotaxime, earning the nickname cefotaximase (CTX). Its debut? A star-studded performance by an Escherichia coli strain in Munich, Germany, in 1989, giving rise to the name CTX-M (M for Munich). Since then, CTXM-type ESBLs have jet-setted their way globally, becoming the hottest ticket in town during the 1990s and the early 2000s. Cue the urgent need for a crash course in CTX-Mology. In the current project, we embarked on a whirlwind, multi-level, in silico analysis to paint a vivid picture of all the natural variants of CTX-M reported to date. Armed with our cyber tools, we delved into six levels of detail: Sequences, Structures, Protein-Drug Complexes, Variant Analysis, Dynamics, and Networking. Our mission is to unravel the mysteries of enzymes of the family beta-lactamases, and CTX-M is a small part of the consortium. We want to pinpoint those pesky mutational sites and assemble a comprehensive database for posterity. This deep dive promises to shed light on the inner workings of CTX-M and bolster our defenses against the dark forces of antimicrobial resistance.
Keywords: Antimicrobial resistance and beta-lactamase, Deepika signg, Hazra lab, Indian Institute of Technology Roorkee
Molecular dynamic simulation study toward improvement of thermostability of nanoluciferase protein for detection of antimicrobial resistance
Adwaita S. R. Nair1 https://orcid.org/0000-0002-3074-1534
1Center for Nanotechnology, Indian Institute of Technology, Departments of
2Physics and 3Bioscience and Biotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Luciferase, a well-known enzyme sourced from the deep-sea prawn Oplophorus, comprises two key components: A catalytic segment of 19 kDa and a non-catalytic segment of 35 kDa. An enhanced variant, Nanoluc, derived from a 16-mutant iteration of the 19 kDa luciferase, was engineered. When activated by furimazine, a coelenterazine derivative, Nanoluc exhibits luminescence that surpasses traditional luciferases such as Fluc and Oluc by a factor of 150. Nanoluc has found widespread utility across various pioneering innovation, research, and development domains. Nonetheless, a deeper comprehension of its structural and dynamic attributes remains imperative. The primary aim of this study was to elucidate how Nanoluc’s unique beta-barrel structure and capped alpha-helices influence its dynamics. Molecular dynamic simulations and assorted analyses were employed to investigate Nanoluc’s thermo-stability across diverse temperature ranges. These analyses encompassed root mean square deviation, distance matrix, dynamic correlation matrix, and radius of gyration. Insights into the unfolding mechanism were gleaned through principal component analysis and free energy landscape assessments, revealing that the beta-barrel core of wild-type Nanoluc unraveled at an accelerated rate. This investigation facilitated a nuanced understanding of the significance of the salt bridge, enabling the identification of new mutations. Comparative analysis demonstrated that introducing salt bridge mutations conferred enhanced thermo-stability and a rigid structure compared to wild-type Nanoluc. By comprehensively scrutinizing the enzyme’s structure and kinetics, superior enzyme variants can be engineered, leveraging this robust system for various research and development endeavors.
Keywords: Nanoluciferase, Luminescence, Molecular dynamic simulation, Sequential resonance energy transfer
Insertion variants in Klebsiella pneumoniae carbapenemase beta-lactamase: A harbinger of subtle turbulence in the realm of antibacterial resistance
Riya Karan1 https://orcid.org/0000-0002-3008-7245
1Department of Biosciences and Bioengineering, Indian Institute of Technology, 2Department of Biosciences and Bioengineering Joint Faculty, Centre of Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) represents a significant global health challenge, highlighting the alarming surge in antibiotic resistance linked to beta-lactamases. Specifically, class A beta-lactamase has shown an unexpected capability to broaden its range of targets through slight amino acid alterations. Nature has intriguingly adapted its strategy for acquiring robust antibiotic resistance by altering and adding amino acids near the active site. Consequently, beta-lactamases can reshape their active site pocket geometry to accommodate diverse medications and develop resistance to a wide array of substrates. Except for ceftazidime-avibactam, Klebsiella pneumoniae carbapenemase (KPC), as a Class A enzyme, can hydrolyze all beta-lactams, combination beta-lactams, and beta-lactamase inhibitors owing to its multiple active site insertions. Understanding the mechanism behind the changes in the KPC enzyme and its interactions with different drugs, facilitated by the insertion of residues around the active site regions, is crucial in the current critical context. Although the insertion does not occur in the recognized catalytic area of class A beta-lactamase, KPC2 exhibits catalytic efficiency about 10 times greater than the investigated insertion variants. Consequently, higher inhibitor concentrations are needed to inhibit this enzyme, compared to most insertion variants requiring lower concentrations. While insertion variants display enhanced binding affinity toward larger substrates, this comes at the cost of reduced enzyme stability. To unravel this enzyme’s behavior and mechanism of action, a comprehensive investigation integrating biophysical, biochemical, kinetic, and biological studies is employed to elucidate the molecular understanding. As antimicrobial resistance poses an increasingly urgent challenge, this holistic approach aims to contribute to developing innovative therapies.
Keywords: Antimicrobial resistance, Antibiotic resistance, Klebsiella pneumoniae carbapenemase, Ceftazidime-avibactam resistance, Insertion variants
Prevalence and outcomes of multidrug-resistant patients in a tertiary care hospital
Ritwick Singla1 https://orcid.org/0009-0005-9520-6697 , Prakhar Sharma1, Kritarth Rawat2, Deepansh Gupta1, Suyash Singh1, Aswathy Sabu1
Department of Pulmonary Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Multidrug-resistant (MDR) organisms are microorganisms, mainly bacteria, that are resistant to two or more classes of antimicrobial agents. There are very few observational cross-sectional studies on the prevalence of MDR pathogens in a Tertiary care hospital. In this study, we analyzed the prevalence of different MDR organisms with their respective outcomes.
Objectives: (i) To estimate the prevalence of different MDR organisms in the respiratory medicine ward and intensive care unit (ICU). (ii) To assess the outcomes of MDR patients in the respiratory medicine ward and ICU.
Methods: For all the patients with a suspected respiratory infection, a lower respiratory tract sample was sent for culture, and the report was analyzed for pattern and prevalence of MDR pathogen in the hospital.
Results: A total of 65 patients were enrolled in the study from September 2023 to January 2023, out of which 39 (60%) patients were successfully treated and were discharged while 26 patients (40%) succumbed to illness. The MDR organisms isolated were Acinetobacter (46.1%), Klebsiella (29.2%), Escherichia coli (12.3%), Pseudomonas (7.6%), and others (4.6%) which includes methicillin-resistant Staphylococcus aureus, Serratia, and Enterococcus. These organisms were treated based on sensitivity patterns and according to recommended guidelines.
Conclusion: The most common MDR organism in the respiratory medicine ward at All India Institute of Medical Sciences Rishikesh was Acinetobacter (46.1%), and mortality was also highest in patients with Acinetobacter infection (46.6%).
Keywords: Multidrug resistant infections, acinetobacter, mortality
Cryptococcal meningitis in an immunocompetent young boy
Jaideep Pilania1 https://orcid.org/0009-0005-9110-7296 , Prasan Kumar Panda2, Anika Malviya1, Ritu Sangwan1
1Department of Internal Medicine (ID Division), All India Institute of Medical Sciences, 2Department of Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Cryptococcal meningitis is an infection caused by the fungus Cryptococcus after it spreads from lungs to the brain and is usually seen in immunocompromised individuals. It is rarely seen in an immunocompetent host. It is usually fatal, if untreated, with high mortality rates. We have encountered an interesting case of cryptococcosis in an immunocompetent young boy. The patient is a 19-year-old male, laborer, occasional smoker, with no prior known comorbidities presented with complaint of acute onset, progressive, diffuse headache with episodes of projectile vomiting and on and off undocumented fever for the past 15–20 days for which he took over the counter medications, with partial relief. He developed an altered sensorium for the last 3–4 days, was admitted to an outside hospital, was given some oral and IV medications, and was referred to a higher center. In Emergency, with the possibility of meningitis, due to the presence of neck rigidity, his lumbar puncture was done, and he was started on empirical ceftriaxone and vancomycin and was admitted to the medicine ward. Fundus examination revealed no papilledema and choroid tubercles. His cerebrospinal fluid (CSF) analysis revealed findings suggestive of bacterial meningitis (likely partially treated) with normal ADA level and CBNAAT negative. Contrast-enhanced magnetic resonance imaging brain revealed leptomeningeal enhancement with vasculitic infarcts. However, due to no significant clinical improvement, his CSF analysis was repeated, whose India Ink came out positive with blood culture having growth of Cryptococcus. He was then treated with amphotericin-B (plain and liposomal as per availability), both intrathecal and IV, and tablet flucytosine, following which he started to improve clinically. This case highlights the importance of identification and early initiation of therapy for cryptococcal meningitis in immunocompetent individual. One should do Indian ink staining of all CSF samples even if patient is immunocompetent. This case signifies the significance of right diagnosis and right treatment in optimizing patient management, reducing AMR risks, and improving clinical outcomes.
Keywords: Bacterial meningitis, Cryptococcosis, Immunocompetent, Indian ink staining
Safety and effectiveness of outpatient parenteral antimicrobial therapy and its role in antimicrobial stewardship – A pilot longitudinal study
Amit Mathur1, Prasan Kumar Panda2 https://orcid.org/0000-0002-3008-7245 , Ravi Kant1, V. S. Pai1, Mukesh Bairwa1, Darab Singh1, Nilanjana1
1Department of Internal Medicine (ID Division), All India Institute of Medical Sciences, Department of Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Outpatient parenteral antimicrobial therapy (OPAT) offers a crucial method for administering IV/IM antimicrobials outside hospitals, enabling patients to complete treatment safely outside and many hospital-acquired events. This pilot study evaluates OPAT’s efficacy, safety, and feasibility in treating infections, emphasizing its role in antimicrobial stewardship. It also addresses the challenges associated with OPAT practice.
Methodology: This pilot longitudinal observational study included patients meeting OPAT checklist criteria and committed to post-discharge follow-up. Pre-discharge education and counseling were provided. Antimicrobial selection depended on infection and patient factors. Daily telephonic monitoring was ensured, and results were documented.
Result: All patients (n = 10) became afebrile, and nine cases saw complete resolution of infections within a month of follow-up and one case after 6 weeks [Figure 1]. No instances of prematurely discontinued OPAT regimens were observed. In addition, no complication related to drugs or vascular line was encountered during close follow-up.
Conclusion: This pilot study highlights OPAT’s 100% safety and effectiveness as an infection management alternative to hospitalization, reducing resource utilization and health-care-associated infections. OPAT directly contributes to integrated antimicrobial stewardship, vital in combating antimicrobial resistance (AMR), aligning with global action plan for AMR in infection prevention and antimicrobial optimal utilization.
Keywords: Antimicrobial resistance, Integrated antimicrobial stewardship, Outpatient parenteral antimicrobial therapy, Telephonic monitoring
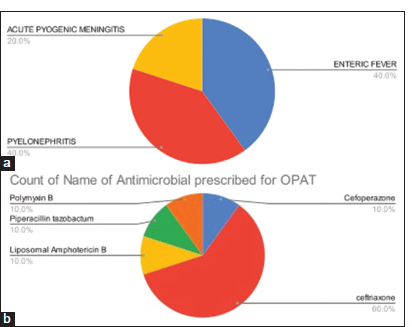
- (a) Pie charts representing infections distribution and (b) antimicrobial distribution.
Prevalence of molecular resistance in multidrug-resistant pathogens: In a tertiary care respiratory intensive care unit
Prakhar Sharma1, Kritarth Rawat1 https://orcid.org/0009-0001-2360-0530 , Ritwick Singla1, Abhishek J. Benur1, Deepansh Gupta1, Suyash Singh1, Aswathy Sabu1
1Department of Pulmonary Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Multidrug-resistant organisms (MDROs) are microorganisms, mainly bacteria, that are resistant to two or more classes of antimicrobial agents. There are very few observational cross-sectional studies on the prevalence of molecular resistance among multidrug-resistant (MDR) pathogens in a tertiary care hospital. In this study, we analyzed the molecular resistance genes and their prevalence in MDR pathogens.
Objectives: (i) To estimate the prevalence of molecular resistance in MDR pathogen in a tertiary care respiratory intensive care unit (RICU).
Methods: For all the patients with suspected nosocomial respiratory infection, multiplex polymerase chain reaction (biofire) was sent from lower respiratory tract sample and report was analyzed for pattern and prevalence of molecular resistance in MDR pathogen detected in tertiary care RICU.
Results: Of the 15 patients enrolled in the study, 11 were male and 4 were female,13 patients showed growth of micro-organism, among them 7/13 showed detection of Acinetobacter, 5/13 showed detection of Escherichia coli, 5/13 showed detection of Pseudomonas aeruginosa, 4/13 showed detection of Klebsiella Pneumoniae, 3/15 showed detection of Rhinovirus, 1/13 showed detection of Staphylococcus aureus, 1/13 showed detection of Serratia marcescens, 1/13 showed detection of Parainfluenza virus, 1/13 showed detection of Streptococcus pneumonia, with prevalence of resistance with gene Cefotaximase-munich CTX-M (8/13), New Delhi metallo-β-lactamase -(10/13), OXA -48 (8/13), VIM – (3/13), IMP (2/13), K. pneumoniae carbapenemase (1/13), Mec-A/C(1/13), MREJ (1/13).
Conclusion: The most common MDR organism in RICU All India Institute of Medical Sciences Rishikesh was Acinetobacter (38%). The most common gene resistance detected was CTX-M (61%).
Keywords: Multidrug resistant infections, biofire, molecular resistance
Study of antimicrobial resistance in blood culture isolates at a tertiary care center in Kumaon region, Uttarakhand
Prateek Gautam1 https://orcid.org/0009-0007-5669-5592 , Umesh1, Shambhavi Singh1, Neha Kaushal1
1Department of Microbiology, Government Medical College, Haldwani, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Bloodstream infection is one of the most important causes of morbidity and mortality globally. This study included 3218 blood culture samples collected from patients suspected of having sepsis and admitted to Dr. Susheela Tiwari Hospital in Haldwani.
Material and Methods: A retrospective analysis of 3218 blood culture samples which were received at the Department of Microbiology, Government Medical College, Haldwani, between January 2023 and December 2023 was performed and evaluated for antimicrobial resistance patterns.
Results: Out of 315 positive cultures, 35.87% were Gram-negative bacteria, with Salmonella enterica subsp. enterica serotype Typhi being the most common, followed by Escherichia coli. Gram-positive cocci accounted for 38.41%, with Coagulase-negative Staphylococcus spp. being the most common. However, pathogenicity could not be ascertained as in most cases, paired blood samples were not received. Fungal isolates accounted for 1.26%. Isolates of Salmonella spp. were most sensitive to third-generation cephalosporins and fluoroquinolones, and E. coli was most sensitive to imipenem and amikacin. Gram-positive isolates showed maximum sensitivity to ciprofloxacin, gentamicin, linezolid, and teicoplanin.
Conclusion: For Gram-positive isolates, linezolid and vancomycin were the drugs of choice, while imipenem and aminoglycosides were the most effective drug for Gram negatives, except for Salmonella spp. for which third-generation cephalosporins and fluoroquinolones were found to be most effective. A unit-based microbiological surveillance, timely and repeated investigation of bacterial flora of bloodstream infection (BSI), monitoring of antibiotic susceptibility patterns, reinforcement of infection control practices, and appropriate isolation and barrier precautions organisms are mandatory to preserve good antibiotic stewardship.
Keywords: Antimicrobial susceptibility, Antimicrobial resistance, Bloodstream infections, Blood culture, Bacterial flora
Clinical profile of multidrug-resistant patients in neurosurgery and trauma ward at a tertiary care hospital – A longitudinal study
Rakhi Yadav1 https://orcid.org/0009-0000-8766-6793 , Sheela Kushwaha1, Bhaskar Sarkar2, Rakesh Kumar Sihag3, Prasan Kumar Panda4, Amber Prasad5, Ajay Kumar2, Deepa Kumari1
1Trauma center, All India Institute of Medical Sciences, Departments of
2Trauma Surgery, 3Neuro Surgery, 4Internal Medicine, 5Microbiology, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: The World Health Organization states that resistant microorganisms (such as viruses, bacteria, fungi, and parasites) may fend off attacks by antimicrobial medications, which results in inefficient treatment that allows diseases to remain and spread. Multidrug resistance (MDR) may potentially be connected to antimicrobial medications. One of the biggest challenges to global public health is antimicrobial resistance (AMR). According to estimates, bacterial AMR caused 4.95 million fatalities worldwide in 2019 and was directly responsible for 1.27 million deaths.
Objectives: (i) To understand the course of illness of MDR-infected patients in Neuro Surgery and Trauma Surgery departments. (ii) To assess the demographical profile and outcome details of MDR patients. (iii) To estimate the compliances to integrated antimicrobial stewardship practices of the health-care workers (HCWs).
Methods: All admitted MDR patients from October 2023 to March 2024 were studied from these two departments. The data of these patients are notified from MDR reporting in live Excel sheet which was further investigated by ground investigators, who document and do random observation in pre-determined format.
Results: Fifty-two patients were studied. Maximum patients, 24 (46.2%) in this study, were in age group 21–40 years. The MDR organisms found among them were Acinobacter Baumannii, Klebsiella Pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and others like Enterococcus, methicillin-resistant coagulase negative staphylococci and majority positive samples were urine [Figure 1a and b]. Nine (17.3%) patient died. Among all HCWs, maximum workers, 32 (61.5%) are vaccinated with routine immunization (Hepatitis B and COVID-19) and 20 (38.5%) are unknowm of their vaccination status.
Conclusion: The MDR mortality rate is observed to be 17.3%. The most common MDR is Acinobacter and Klebsiella in these two department patients. Still, 1/3rd HCWs are not vaccinated or with unknown status who give care to these MDR patients.
Keywords: Antimicrobial resistance, Antimicrobial stewardship, Healthcare worker vaccination
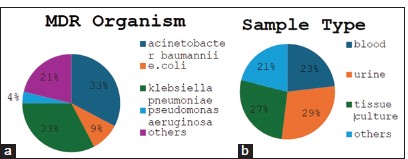
- Pie chart distribution of (a) Multi drug resistant organism and (b) sample types.
Exploring the novel beta-lactamase ElBla2 as a gateway to deciphering the mystery of New Delhi metallo-beta-lactamase
Jyoti Barman1, Saugata Hazra1,2 https://orcid.org/0000-0002-3074-1534
1Department of Biosciences and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The New Delhi metallo-β-lactamase (NDM) poses a pressing threat to public health, evident from its rapid global dissemination since its initial identification in 2009. Diverging notably from other Metallo-β-lactamases (MBLs), NDM has spurred researchers to seek its close relatives, aiming for insights crucial to inhibitor design. ElBla2, sourced from the marine bacterium Erythrobacter litoralis HTCC 2594, shares sequence homology with NDM. Remarkably, ElBla2 exhibits a higher amino acid sequence resemblance to NDM-1 (56%) than any previously reported MBLs. Enzymatic assays and secondary structure alignment corroborate the substantial similarity between these enzymes. Whole genome comparisons among four Erythrobacter species reveal highly conserved genes flanking elbla2, hinting at its potential loss during evolutionary processes. In addition, predictive analyses unveil two prophages, 13 genomic islands, and 25 open reading frames associated with insertion sequences in E. litoralis HTCC 2594’s genome. Unlike NDM-1, however, the chromosome-encoded ElBla2 is not situated within or proximal to these mobile genetic elements, suggesting limited inter-strain transferability. Following phylogenetic analysis, researchers propose reclassifying E. litoralis HTCC 2594 as a novel species: Erythrobacter spp. HTCC 2594. The secondary protein structures of ElBla2 and NDM-1 exhibit striking similarities, each comprising 13 β-sheets and six β-helices. Numerous active sites found in NDM-1 are conserved in ElBla2, including N220, pivotal for lactam carbonyl group interaction, and K211, facilitating the orientation of the negatively charged carboxylate typical in β-lactam substrates. Despite these parallels, differences exist in catalytic efficiencies between the two enzymes. Notably, while ElBla2 displays low Km values akin to NDM-1 for most cephalosporins, its turnover rate (kcat) for various substrates falls below that of NDM-1. Specifically, ElBla2 demonstrates its highest catalytic efficiency with meropenem, although at a lower turnover rate compared to NDM-1. Full steady-state kinetics profiling and molecular dynamics simulations of loop regions surrounding the active sites were conducted to gain deeper insights into these distinctions.
Keywords: ElBla2, New Delhi metallo-β-lactamase, Beta lactam drugs, Catalytic efficiencies
Antifungal resistance and new strategies to control fungal infections
Sheetal Kumari1, Anjali, Mayank1, Brijesh1, Neha Chauhan2 https://orcid.org/0000-0002-1687-0831
Departments of 1Medical Laboratory Technology and 2Microbiology, School of Paramedical and Allied Health Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Despite improvements in antifungal medicines over the previous 30 years, antifungal resistance remains a major concern in clinical practice. In the past 10 years, the molecular mechanisms behind this phenomena have been widely studied. Following a quick introduction of currently available antifungals, this paper will go into detail on the molecular underpinnings underlying antifungal resistance. Major resistance pathways appear to be essential as a result of antifungal resistance effector gene dysregulation. This dysregulation is the result of point mutations in transcriptional regulators of these effector genes. Resistance can also arise as a result of point mutations in genes that code for antifungal targets. In addition, we further detail novel tactics now conducted to explore alternative therapeutic targets and antifungals. Screening collections of natural or synthesized chemical compounds is the primary method for identifying novel antifungals. Genome-wide techniques are used to discover novel potential antifungal targets and gain a better knowledge of the biology of human pathogenic fungi.
Keywords: Antifungal resistance, Point mutation, antifungal targets, Gene dysregulation pathogenic fungi
Role of Klebsiella pneumoniae carbapenemase variant 107 in rendering anti-microbial resistance in Klebsiella pneumoniae
Adarsh Singh1, Riya Karan1, Saugata Hazra1 https://orcid.org/0000-0002-3074-1534
1Department of Biosciences and Bioengineering, Indian Institute of Technology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Beta-lactam antibiotics remain the most widely used antibiotic in human history. However, their efficacy has been challenged by the expression of beta-lactamases that are able to inactivate all beta-lactam drugs including carbapenems. Among beta-lactamases, Class A enzymes are majorly responsible for developing resistance in Gram-negative bacteria, including ESKAPE pathogens, which are highly associated with hospital-acquired infections. Klebsiella pneumoniae carbapenemase (KPC) Class A carbapenemase produced by K. pneumoniae is inhibited by combinatorial therapy like ceftazidime-avibactam, like all other potent carbapenemases. KPC-107 is one of the insertional mutants of the wild type, KPC-2, has 28 amino acid insertion near the omega loop region which changes the dynamics of the active site. Despite this huge insertion, the enzyme was successfully over-expressed and purified. The enzymatic efficiency (kcat/Km) of KPC-107 and KPC-2 against nitrocefin showed a drop of 6.6-fold and lower efficiency against most known substrates even though it shows higher binding constant with bulkier substrates. KPC-107 also exhibits 6-fold decrease in resistance against diazabicyclooctanes as compared to KPC-2. In addition, the optimum temperature of activity of KPC-107 peaks around 30°C, far below than that of KPC-2 which is around 40°C indicating a decrease in temperature stability of the enzyme as expected from such a big insertion. The selection pressure and advantage of this supposed structurally destabilizing and functionally deteriorating mutation and its consequences on global AMR is a complete mystery. To unravel the reason behind the existence of KPC-107 and the threat it poses to our fight against AMR, further biophysical and structural analysis needs to be done.
Keywords: Klebsiella pneumoniae carbapenemase, Klebsiella pneumoniae carbapenemase -107, Class A beta-lactamase, Carbapenemases
Identification and antimicrobial susceptibility infection patterns of various Gram-negative bacilli isolated from blood culture in patients with bloodstream infection
Harshita Chandra1, Arpana Singh1, Barnali Kakati1 https://orcid.org/0000-0002-1592-9109
1Department of Microbiology, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Background: Gram-negative bacteria are among the world’s most significant public health problems due to their high resistance to antibiotics. These microorganisms have significant clinical importance in hospitals because they often require patients to be in the intensive care unit and patients are at high risk of morbidity and mortality. Blood culture remains the main diagnostic method for appropriate treatment in hospital-acquired bloodstream infections and to diagnose any bacteremia in patients. The present study was therefore done to identify various Gram-negative bacteria isolated in blood culture and to determine their antimicrobial susceptibility pattern.
Material and Methods: A retrospective study was done over a period of 3 months from November 2023 to January 2024 in the Bacteriology section of the Microbiology department, where different Gram-negative bacteria isolated on blood culture were noted. The antibiotics with their resistance patterns and minimum inhibitory concentration (MIC) were also recorded.
Results: A total of 180 patients were included in the study, with a male-to-female ratio of 1.7 and a mean age of 28 years. It was observed that the most common Gram-negative bacteria isolated from blood culture was Klebsiella pneumoniae, followed by Escherichia coli. The distribution showed a higher prevalence of Acinetobacter baumannii in the intensive care unit and K. pneumoniae in the non-intensive care units. The analysis of antibiotic resistance showed that K. pneumoniae was most susceptible to tigecycline with MIC of ≤0.5 and was resistant to amoxicillin, tazobactum, cefuroxime, cefoperazone, cefepime, ertapenem, imipenem, moropenem, amikacin, ciprofloxacin, trimethoprim, levoflox, minocycline, gentamicin, and ceftazidime.
Conclusion: In our setup , K. pneumoniae is the most commonly isolated Gram-negative bacteria which was multidrug resistant. This addresses the fact that infection control practices should be strictly followed. Regular monitoring should be done and this antibiogram will help to start the appropriate treatment.
Keywords: Gram-negative bacteria prevalence in blood culture, Antimicrobial sensitivity, Minimum inhibitory concentration, Klebsiella pneumoniae, Escherichia coli, Tigecycline, Colistin
Determination of antibiotic-resistant bacteria antimicrobial resistance in organic vermicompost
Alaa Eddin Alhmeidi Alkhatib1 https://orcid.org/0009-0003-6103-014X , Rajsekhar Adhikary1, Saugata Hazra1
Department of Biosciences and Bioengineering Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Vermicompost, which is produced by earthworms and used to enhance soil quality in agriculture, is suspected to contain antimicrobial-resistant bacteria due to the animal and plant waste it’s composed of due to the widespread use of antibiotics in animal farming. To determine the antibiotic resistance in vermicompost, the antibiotic resistance profiles were assessed using the replica plating method of fifteen antibiotics, and the microbial composition of vermicompost samples was assessed from thirty samples from twelve states across India as an initial step. Next, based on the higher resistance loads, 16S ribosomal ribonucleic acid next-generation sequencing was performed on nineteen samples. Among the antibiotics tested, the bacteria showed the highest resistance levels with aztreonam, cefixime, and cefuroxime, while ofloxacin, meropenem, and doripenem showed the lowest resistance. Intriguingly, two samples from Rajasthan showed resistance to all antibiotics, while another eight showed resistance to fourteen antibiotics. Through the 16S metagenomic approach using the sequence of the V3 region, the genera Pseudomonas, Bacillus, Clostridium, and Rahnella under the phyla Proteobacteria were found to be the most abundant genera present in all samples. Alpha diversity analysis revealed varying degrees of diversity in the sampled areas. Simpson’s diversity index (D) was lowest in Beng-02 and Beng-01, while it was highest in UP-07 and MP-01. Furthermore, the community structure analysis showed a significant divergence across the communities under consideration.
Keywords: Vermicompost, Antibiotic resistance, Antimicrobial resistance, 16S rRNA sequencing, Next-generation sequencing
Antimicrobial resistance: Conventional diagnostics system tools
Anjali1, Mayank Purohit1, Sheetal Sharma1, Brijesh Kumar1, Neha Chauhan2 https://orcid.org/0000-0002-1687-0831
Departments of 1Medical Laboratory Technology and 2Microbiology, School of Paramedical and Allied Health Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) is one of the most serious risks to public health. Hence, there is an increasing demand for methodologies and technology that allow for quick antimicrobial susceptibility testing (AST). Conventional methods and technology for AMR diagnosis and AST in clinical microbiology are time-consuming, have long turnaround times, and are typically expensive. As a result, empirical antibiotic medicines are administered, which promotes AMR spread, resulting in higher mortality rates and higher health-care costs. Antimicrobial susceptibility testing is a critical step in identifying effective antimicrobial medicines to treat infectious illnesses. The procedures employed in diagnostic microbiology laboratories are constantly evolving. There is a growing emphasis on automation, genotypic, and micro/nanotechnology-based advances. Automation in detecting systems, as well as the integration of computers for online data processing and data exchange, represent significant advances in the versatility of automated approaches currently in use. Genotypic methods use molecular amplification and genome sequencing to identify a specific genetic marker linked with resistant phenotypes.
Keywords: Antimicrobial resistance, Antimicrobial susceptibility testing, Turnaround times, Genotypic methods, Micro/nanotechnology-based techniques.
“Unveiling boronate-based transition state analogs: A journey from tankana kshara to next-generation antibiotics” exploring the wisdom of vedic microbiology in crafting novel therapeutics against beta-lactamase enzymes
Madhusmita Tripathy1, V. Venkatesh2,3, Kunal Dhankar1, Saugata Hazra1, 2
1Department of Bioscience and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology,
3Department of Chemistry, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The peril of antibacterial resistance: Pioneering boronate-based solutions antibacterial resistance looms as a formidable clinical menace to global health, propelled by the rampant overuse of antibiotics. Pathogens adeptly mutate in response to repeated antibiotic exposure, rendering traditional treatments ineffective. In regions like India, where grassroots awareness of proper hygiene and medication adherence remains nascent, innovative strategies are imperative to combat the rise of drug-resistant pathogens. Among the arsenal of antibacterial therapeutics, penicillin and cephalosporins, revered for their efficacy in inhibiting bacterial cell wall biosynthesis through transpeptidase inhibition, are pillars of clinical prescription. However, a primary strategy bacteria employ to resist the onslaught of beta-lactam antibiotics is the production of beta-lactamases. These enzymes effectively dismantle the structural integrity of antibiotics, conferring resistance. Drawing inspiration from ancient Ayurvedic remedies, particularly Tankana Bhasma, historically employed for various ailments, including ulcers, asthma, coughs, and urinary tract infections, the Hazra Group embarks on a modern scientific quest. Tankana Bhasma, chemically identified as borax, serves as the foundation for boronate, a potential lead compound in mimicking carbon as a transition state analog of beta-lactamase inhibitors. In our ongoing endeavor, we unveil a multidisciplinary approach to unravel the intricacies of beta-lactamases and forge ahead with the development of boronate-based transition state mimics, heralding a new era of therapeutics resistant to drug resistance. Our presentation showcases promising results, demonstrating the efficacy of our newly designed boronate-based transition state analogs, poised to revive the efficacy of early-generation penicillin and cephalosporin antibiotics through innovative combinatorial strategies.
Keywords: Beta-lactamases, Boron chemistry, Boronate-based transition state analogs, Antibacterial resistance
Unveiling the hidden antimicrobial treasures: Actinobacteria associated with leaf cutter ants of Uttarakhand
Megha Choudhary1 https://orcid.org/0000-0002-6068-5983 , Vijay Kumar1, Bindu Naik2, Vivek Kumar1, Akhilesh Kumar1
1Himalayan School of Biosciences, Swami Rama Himalayan University,
2Department of Food Science and Technology, Graphic Era University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) remains a critical global health threat, with bacteria, viruses, parasites, and fungi increasingly becoming resistant to existing medications. Urgent action is needed to address this crisis, to prevent regressing to a pre-antibiotic era. Recently, insects have been viewed as a vast and largely untapped reservoir of unique microorganisms possessing antimicrobial potential. Insects such as ants, termites, beetles, wasps, honeybees, and grasshoppers have been identified as carriers of insect-associated actinobacteria, predominantly Streptomyces and Pseudonocardia species. Despite this, our understanding of the chemical ecology surrounding insect-associated actinobacteria remains limited. In this context, 24 samples of leaf cutter ants were collected from various regions of Uttarakhand; from which, 20 actinobacterial isolates were recovered belonging to five different genera based on polyphasic taxonomy. Screening was done against the drug-resistant fungal pathogens including Candida albicans (MTCC 183), Candida parapisilosis (NCIM 3082), Candida tropicalis (NCIM 3661), Microsporum canis (MTCC 2820), Trichophyton rubrum (MTCC 296), Aspergillus fumigatus (MTCC 2544), Aspergillus flavus (MTCC 1883), and Aspergillus niger (MTCC 281). Six (30%) of the isolates exhibited different ranges of antifungal activity. High-resolution mass spectrometry analysis of crude extracts of the most promising isolate (Streptomyces spp. AJ-1) detected the presence of previously known compounds with antifungal activity (like antimycin) along with a few potentially novel bioactive compounds while, whole-genome analysis (antiSMASH, BAGEL4) further confirmed these results. The results of this study highlight the enormous potential of microorganisms associated with ants to produce novel compounds which may help to combat AMR.
Keywords: Antimicrobial resistance, Actinobacteria, Leafcutter ants, Antifungal, AntiSMASH
The use of statistical data to graph anti-microbial resistance
Aditya Pratap Singh1 https://orcid.org/0000-0002-1592-9109
1Department of Microbiology, Himalayan Institute of Medical Sciences Swami Rama Himalayan University, Jolly Grant, Dehradun, India
E-mail: nareshchettri15@gmail.com
Anti-microbial resistance is an issue of global concern. The bacteria, viruses, and fungi have been able to evolve hand in hand with the new strategies of antibiotic regimen. This has led to the development of exploding anti-microbial resistance. The use of data can be an important aid in understanding the statistical pathogenesis and distribution of diseases in society. Apotheek, which translates to pharmacy in the Netherlands, is a computerized medicine dispensing booth. The physician writes a prescription, and the diagnosis and prescription are emailed to the patient. The Apotheek machine dispenses the medicine with an instruction to use the medicine. In the Indian Scenario, the same thing could be tested by this. The controlling authority can know about the prescription written and the amount of antibiotics dispensed to the patient. This prevents mindless prescription writing and lets us know about the response. For example, the case of tuberculosis. A rise in rifampicin resistance cases has been a consequence of clinical transmission in society. India, being a populous country, allows the resistance to develop at a faster rate. A data-strong compendium of cases and their demographic distribution can help us identify hotspots of disease and patient’s response to treatment. This will also help in a more careful prescription and judicious use of these miraculous drugs, a new term of Microlife Sanctuary.
Keywords: Anti-microbial resistance, Statistical pathogenesis, Apotheek, Rifampicin resistant, Hotspots, Microlife sanctuary
To study Sokal, Hasford, EUTOS, and ELTS prognostic score in predicting hematological remission and molecular response in chronic myeloid leukemia patient
Randhir Kumar Mahato1 https://orcid.org/0009-0002-9511-6518 , Nadia Shirazi1
1Department of Pathology, Himalayan Institute of Medical Sciences Swami Rama Himalayan University, Jolly Grant, Dehradun, India.
E-mail: nareshchettri15@gmail.com
The primary goal of chronic myeloid leukemia (CML) management is to stratify patient’s risk to identify the most optimal therapeutic regimen. The Sokal, Hasford, and EUTOS risk ratings were developed to predict patients on treatment.
Aim: To perform a comparative study of CML prognostic indicators (Sokal, Hasford, and ELTS) at Swami Rama Himalayan University CML-CP patients with their demographical and hematological parameters.
Methods: This is a retrospective study performed on 71 Ph+ CML-CP patients who were before and after being administered imatinib orally and study their demographical and hematological data. 30/71 were females and 41/71 were males with median age 38 years (range 18–75 years). 3 (4.22%), 27 (38.02%), and 41 (57.75) patients were discriminated into low, intermediate, and high-risk Sokal score, respectively. 12 (16.90%), 37 (52.11%), and 22 (30.99%) patients were discriminated into low, intermediate, and high risk of Hasford score, respectively, and 6 (8.45%), 28 (39.44%), and 38 (53.52%) were patients divided into low, intermediate, and high, respectively. Major molecular remission (MMR) was present 15 (21.13%), and 15 (21.13%) absent in female. In male, MMR was 14 (19.72%) present and 27 (38.03%) present, highly present, and absent in age group range is 20–40 years of patients.
Conclusion: The study found that Sokal and EUTOS significantly predict treatment outcomes for CML-CP patients taking imatinib, and patients aged 20–39 are highly effective.
Keywords: Chronic myeloid leukemia (prognosis, Chronic myeloid leukemia), Sokal, Hasford, EUTOS
Interdisciplinary approach to combat anti-microbial resistance
Aditya Pratap Singh1 https://orcid.org/0000-0002-1592-9109
1Department of Microbiology, Himalayan Institute of Medical Sciences Swami Rama Himalayan University, Jolly Grant, Dehradun, India
E-mail: nareshchettri15@gmail.com
The present decade has seen a global pandemic COVID19, a consequence of genetic mutation of virus which caused a global havoc. The disease rendered many allopathic treatments useless. A united global effort developed the vaccine to overcome this resistance. Hence, the antimicrobial resistance has been on the rise since then and troubled the health-care providers worldwide. The general public has understood that although allopathic medicines are irreplaceable but are limited too by antimicrobial resistance. Hence, a holistic approach of health is to be addressed. Inter-disciplinary approach can be an alternative. The complete dependence on allopathic medicine can be relaxed by switching on to some extent to Ayurveda, Yoga, Unani, Siddha, and Homeopathy. The Indian traditional sciences can be an alternative for some if not all cases. An example of Tu Youyou’s discovery of Artemisinin due to an extensive support framework for Chinese traditional medicine by Chinese government. The old medicine of Ayurveda too is unique in its nature. The example of storing water in copper utensils, eliminates the diarrhea causing Escherichia coli, the use of turmeric in wound healing and the use of nanoparticles in Bhasma can also be utilized. The Ayurvedic medicine is miraculously biocompatible, for example, Bhasma is usually consumed with ghee or honey these substances have better biocompatibility. Turmeric derived from the rhizome has been able to scavenge free radicals and superoxides. The major disadvantage of Ayurveda medicines is for example bhasma is contaminated by metallic particles, turmeric has shorter life span and a lack of research in the field. Hence, a need arises to divide the burden of treatment amongst other alternative medicine to decrease our dependence on Western medicine. An Indian version of fusion medicine could be a contribution to the world of medicine by India.
Keywords: Antimicrobial resistance, Inter-disciplinary approach, Tu youyou, Copper utensils, Nanoparticle, Biocompatability, Turmeric
Environmental BL tester: Tools for detection and surveillance of environmental antimicrobial resistance
Ankit Kumar singh1, Rajsekhar Adhikary1, Saugata Hazra1,2 https://orcid.org/0000-0002-3074-1534
1Department of Biosciences and Bioengineering, Indian Institute of Technology, Roorkee, Uttarakhand, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antimicrobial resistance (AMR) not only impacts human health but also jeopardizes the fundamental pillars of a country’s or continent’s economic status by causing harm in mainly four domains: Human, animal, food, and environment. Environmental samples, including wastewater, soil, and river/lake water, serve as significant sources for the escalating emergence of drug-resistant bacteria due to improper discharge. These bacteria can enter the environment and, over time, pose serious health risks. It is imperative to recognize the gravity of AMR, which is spreading rapidly. The primary aim behind developing the environmental BL-tester is to assess the AMR scenario in various environmental locations, addressing the insufficient data from many parts of India to form a comprehensive understanding and establish global correlations. In this study, a total of 12 samples were collected from different areas of Roorkee town, Haridwar, India focusing on drainage, Solani river, Ganga River, and Dairy. To estimate the total drug-resistant bacterial load, we followed the standard methods and protocols (Clinical and Laboratory Standards Institute 2022), using standard agar and broth dilution methods. Drug resistance bacterial screening was performed using meropenom, oxacillin, cephalothin, ceftazidime, colistin, kanamycin, ofloxacin, and trimethoprim. All the samples showed high level of antibiotic resistant to the above antibiotics except for ofloxacin and meropenem. These two antibiotics showed the lowest level of resistance compared to the others. The growth pattern suggests the presence of active drug-resistant bacteria in samples. To detect the presence of enzyme-mediated drug resistance, a dye named Nitrocefin was used. It has a property of chromogenic shift (390 –486 nm) visible to the naked eye. This environmental BL-Tester provides a highly specific and fast colorimetric assay for detecting the presence of AMR in a sample and also helps with regular surveillance of environmental AMR with a specific chromogenic reaction within a few minutes (10–15 min).
Keywords: Antibiotics, Antimicrobial resistance, Chromogenic assessment, Drug-resistant bacteria, Environmental BL-tester
Exploring sequence, structure, and dynamics: A comparative study of intercellular adhesion operon proteins involved in biofilm formation in clinically isolated Staphylococcus species
Aaryan Bhardwaj1, Rajsekhar Adhikary1, Saugata Hazra1,2 https://orcid.org/0000-0002-3074-1534
1Department of Biosciences and Bioengineering, Indian Institute of Technology, 2Centre For Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
A biofilm is a complex community of bacteria that forms on diverse surfaces, enveloped in a self-produced polymeric substance. This biofilm enhances virulence and aids in adherence, colonization, immune evasion, and resistance to antimicrobial therapies. Biofilm development primarily relies on the creation of an extracellular matrix binding the constituent cells together. Key exopolysaccharides such as polysaccharide intercellular adhesin (PIA) or poly-N-acetylglucosamine play a pivotal role in biofilm accumulation and maturation. This matrix comprises PIA, extracellular deoxyribonucleic acid, and surface proteins (cell wall-anchored proteins). Biofilm formation is highly coordinated through intra and intercellular signaling networks within the bacterial community. In Staphylococcus species, PIA production is governed by the intercellular adhesion (ica) operon, housing genes icaA, icaD, icaB, and icaC. This study focuses on the biofilm formation patterns of three clinical Staphylococcus isolates (Staphylococcus aureus CHRFS5, Staphylococcus hominis S19, and Staphylococcus epidermidis S48B) and delves into the structural variations within the ica operon among them. The whole genome sequences of these three clinical isolates exhibit distinct variations in their genetic composition and operon structure. Specifically, S. aureus CHRFS5 and S. epidermidis S48B carry all four genes of the operon, including the icaR gene, whereas S. hominis S19 possesses icaA, icaB, and icaC genes. This structural diversity assessment among the ica isoforms serves as a genetic tool for species differentiation and a marker for virulence in clinical treatment and the development of antimicrobial resistance.
Keywords: Biofilm, Antimicrobial therapy, operon
Assessment of antibacterial resistance in clinical bacteria using a competitive BL tester
Neha Bharti1 Rajsekhar Adhikary1, Saugata Hazra1,2 https://orcid.org/0000-0002-3074-1534
1Department of Bioscience and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The emergence of antibiotic-resistant bacteria poses a significant threat to human health. This study describes a methodology for diagnosis antibacterial resistance in clinical bacterial isolates using a competitive BL tester, a rapid and efficient microdilution technique. This methodology outlines the culturing of clinical isolates, preparation of antibiotic and bacterial dilutions, and subsequent incubation steps in a microplate. Competitive BL testers utilize nitrocefin, a chromogenic β-lactam substrate, to assess bacterial growth and β-lactamase activity. Bacterial viability and potential resistance to antibiotics are inferred based on nitrocefin cleavage by β-lactamase enzymes, resulting in a colorimetric change. Nitrocefin addition followed by absorbance measurement allows for the determination of the lowest antibiotic concentration that inhibits bacterial growth. Established breakpoints are then used to categorize the bacteria as susceptible, intermediate, or resistant to the tested antibiotics. This method provides a consistent and trustworthy way to test how well antibiotics work against bacteria in hospitals and clinics. This helps doctors choose the right antibiotics, which can stop the spread of drug-resistant germs. Higher absorbance indicates bacterial growth and β-lactamase activity. Conversely, lower values indicate inhibited bacterial growth and reduced β-lactamase activity.
Keywords: Antimicrobial resistance, bacterial growth, Beta-lactamase activity
Isolation and characterization of multidrug resistance gram-negative bacterial isolates collected from tertiary care hospital in Dehradun and their treatment using artemisia vulgaris extracts
Shilpa kukreti1, Neha Chauhan2 https://orcid.org/0000-0002-1687-0831
1Shri Guru Ram Rai University, 2Department of Microbiology, School of Paramedical and Allied Health Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Multidrug-resistant (MDR) microorganisms are becoming a serious problem today. Due to rapid development of multidrug resistance against chemotherapeutic medicines, primarily antibiotics screening for potential antimicrobial effects in diverse medicinal plants, such as herbs, has become crucial. These medicinal drugs must be accessible, safe, affordable, and effective. The goal of this investigation was to determine the medicinal potency of crude plant extracts of Artemisia vulgaris in inhibiting MDR gram negative (GN) enteric bacterial isolates. Based on an increase in polarity to inhibit the GN enteric bacterial isolates, seven distinct crude extracts (hexane, petroleum ether, chloroform, acetone, ethyl acetate, methanol, and aqueous) were extracted using the Soxhlet extraction method. Various quantities of the extracts were obtained by dissolving them in Dimethyl Sulfoxide. Macro broth dilution test minimum inhibitory concentration and Agar well diffusion procedure were employed to evaluate the sensitivity of MDR GN bacteria isolates against crude plant extracts. All of the extracts were poured on 24 well microtiter plates as two-fold serial dilutions using Mueller-Hinton Broth (Hi-media, Mumbai, India) as diluents, based on the Agar well diffusion test results. According to the findings, plant extracts significantly inhibited several isolates of MDR GN enteric bacteria. Researchers studying the effects of different infections and developing new drugs may find new hope in this regard.
Keywords: Gram negative enteric bacteria, Dimethyl sulfoxide, Minimum inhibitory concentration, Multidrug-resistant
Approach to multidrug-resistant patients in the medicine ward and intensive care unit at a tertiary care hospital- A longitudinal study
Vinita Saini1, Prasan Kumar Panda2 https://orcid.org/0000-0002-3008-7245 , Mukesh Bairwa1, Deepa Kumari1, Amber Prasad1, Vanya Singh1
1Department of Medicine, Microbiology, and Nursing, All India Institute of Medical Sciences, 2Department of Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Nowadays, many antimicrobials are being taken without doctor’s advice and multiple drugs are used to treat patients, hence pathogenic bacteria start growing in their bodies and generate multidrug resistance (MDR). One of the biggest challenges to global public health is antimicrobial resistance (AMR). According to estimates, bacterial AMR caused 1.27 million deaths worldwide. Hence, a global action plan was made to stop this AMR pandemic.
Objectives: (i) To understand the course of illness of MDR-infected patients in Medicine department. (ii) To estimate the recovery and death rate of MDR patients. (iii) To estimate the compliances to integrated antimicrobial stewardship practices of the health-care worker (HCWs).
Methods: All admitted MDR patients from May 2023 to March 2024 were studied. The data of these patients are notified from MDR reporting in live Excel sheet which was further investigated by ground investigators, who document and do random observations in pre-determined format. After collecting the data from the Excel sheet, the ground investigator went to the patient area and crosschecked the patient status, checked the sample accuracy by correlation of symptoms, and ensured that the patient was isolated or contact precautions would be done by HCW. A WhatsApp group was also created for the accuracy of the data, in which all the ground investigators kept updating the live data of the patients of their respective areas (such as how many patients recovered and were discharged and how many patients died) to their area investigator weekly.
Results: Total number of deaths/discharged/DOR/LAMA/admitted patients from total MDR patient data of medicine ward and intensive care unit (ICU) is described [Figure 1a]. The MDR organisms isolated were Klebsiella, Escherichia coli, Acinetobacter, Enterococcus, Pseudomonas, and others out of 295 MDR patients [Figure 1b]. Klebsiella and Acinetobacter were the most common MDRs in blood samples while E. coli and Enterococcus were the most common MDRs in urine samples. Only 1/3rd MDR patients could be isolated till sterilization of the samples/discharged. HCWs doing duty in these patients were vaccinated 86% for hepatitis B and coronavirus disease 2019.
Conclusion: The MDR mortality rate is observed to be 29.1%. The majority of MDR patients were affected by Klebsiella, E. coli, and Acinetobacter in the medicine ward and ICU. MDR patients were isolated in 1/3rd cases and HCW vaccination rate was 86%.
Keywords: Antimicrobial resistance, Antimicrobial stewardship, Multidrug resistance isolation, Health-care worker vaccination
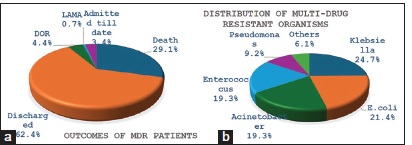
- Pie chart distribution of multidrug resistance organism’s (a) outcome and (b) types.
Deciphering the sequence, structure, and dynamic variations of deoxyribonucleic acid- binding cox protein in P2 and P2-like enteric bacteriophages: advancing phage based alternative anti-microbial resistance therapeutics through in silico and in vitro approaches
Mousumi Hazra1 https://orcid.org/0000-0001-5791-7993 , Kunal Dhankhar2, Saugata Hazra2 https://orcid.org/0000-0002-3074-1534 , Ramesh Chandra Dubey1
1Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar, 2Department of Biosciences and Bioengineering, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The Cox protein is pivotal in determining the transition between lytic and lysogenic phases in enteric bacteriophages. It is a crucial candidate for developing phage-based treatments against drug-resistant pathogens - a pressing global concern amid rising antimicrobial resistance. Cox binds to deoxyribonucleic acid (DNA) as a regulatory protein, promoting the lytic phase. Its primary function revolves around DNA interaction, which governs the expression of various promoters. Our recent research extensively investigated the sequence, structure, and dynamics of the Cox protein and its DNA binding. The scarcity of readily available information posed a significant challenge, prompting us to conduct a thorough literature review to compile a comprehensive Cox protein database enriched with relevant details. Leveraging this database alongside sequence-based phylogenetic and conservation analyses, homology modeling, atomic-level docking, and molecular dynamics simulations, we gained valuable insights into the sequence-structural dynamics crucial for functional comprehension. Furthermore, we successfully cloned, overexpressed, and purified two Cox proteins from Escherichia coli and Salmonella phages. The purified protein enabled us to characterize Cox proteins using biophysical and biochemical techniques. A protein-DNA interaction study used designed promoter DNAs to validate the simulation results. This effort provided valuable mechanistic insights into the protein’s function. Overall, our study holds promise in elucidating the functional mechanisms of the Cox protein, thereby influencing lytic-lysogenic decisions, and contributing to the design of engineered phage-based therapeutics.
Keywords: Cox, Deoxyribonucleic acid-binding protein, Homology modeling, Docking, Molecular dynamics simulation, Phage therapeutics
Combating antimicrobial resistance through SNS and metal-doped nanoparticles: A perspective study
Kanishka Sharma1, Rajsekhar Adhikary1, Deepika Singh1,2, Saugata Hazra1,2 https://orcid.org/0000-0002-3074-1534
1Department of Bioscience and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Antibiotics resistance (AMR) is a critical challenge in global public health concern with forecasts a death of nearly 10 million by 2050 due to inefficacy of current antibiotics toward resistance. In search of alternate solution, nanoparticle is one of the potential substitutes. In this study, Photoinductive reactive oxygen species (ROS) generating sulphide nanoparticles (SNS), reactive oxygen species (ROS) and metal-doped nanoparticles possess potent antibacterial efficacy with specific mechanism of action. Preliminary characterization of the nanoparticles was done by ultraviolet (UV) spectra, absorption band gap determination, scanning electron microscopy (SEM), and ROS generation efficiency. The antibacterial efficacy was assessed by minimum inhibitory concentration (MIC) determination, scanning electron and fluorescence microscopic study, and cell membrane damage study of two biofilm forming pathogens Staphylococcus aureus and Pseudomonas aeruginosa. In UV spectroscopy analysis identified a prominent absorption peak at 400 nm for SNS nanoparticle and this characteristic wavelength is also observed in metal-doped nanoparticles, an indicate a band gap of approximately 3.5 eV for SNS nanoparticle and this value remain consistent for metal-doped nanoparticle aligning with the band gap of pure SNS, In MIC value decrease after photoinduction, it could a change in microbial activity or population due to the induction process or ROS generation, During SEM analysis nanoparticle cause damage to cell wall or cell membrane due to their interaction with the surface of microorganisms (Staphylococcus and Pseudomonas) this damage may manifest as disruption or rupture the cell wall, In biofilm exerts stress on the cell membrane disrupting its structure and functionality it leading to compromised cellular integrity and in fluorescence microscopy was used to antibacterial effect of SNS nanoparticle to the microorganism like they were affected by the presence of SNS nanoparticle their highlighting areas of bacterial cell death and damage the cell membrane that caused by the nanoparticles.
Keywords: ROS generation, Nanoparticles, fluorescence microscopy
Unraveling the structural and functional aspects of a novel metallo hydrolase from Peptoclostridium difficile toward understanding new anti-microbial resistance emergence
Abirlal Mukherjee1, Saugata Hazra1 https://orcid.org/0000-0002-3074-1534
1Department of Biosciences and Bioengineering, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Peptoclostridium difficile infection (PDI) is a common reason behind nosocomial diarrhea in humans and neonatal diarrhea in wild and domestic animals worldwide. In 2018 class D β- lactamase was found in the pathogen that causes PDI. Our investigation finds the presence of another novel metallo hydrolase in the same organism. The major objective of this work was to evaluate the structure function relationship of this new enzyme. Cloning and cell viability assay were performed. Subsequently, the enzyme was overexpressed and purified. Elaborated steady-state kinetics and metal characterization were conducted using (MP-AES), electron paramagnetic resonance, and ultraviolet– visible spectroscopy. Minimum inhibitory concentrations of different β-lactams were derived. Failure in crystallization of such a new protein led us to generate the AlphaFold model and analyze in silico receptor-ligand interactions using docking. Two Zn2+ ions were found to be present in the enzyme active site with tetrahedral and trigonal bipyramidal coordinations but the active site residues were different from Metallo-Beta-Lactamases (MBL). Aztreonam was hydrolyzed by the enzyme unlike MBL, beside different penicillins, cephalosporins, and carbapenems. Esterase activity was also found in this enzyme. The enzyme had very low ability to bind other transition metals in its active site. Our work classifies this enzyme in metallo hydrolase family by differentiating from all MBL subclasses B1, B2, and B3. Probable active site of the novel biocatalyst is identified. Indeed, this study identifies and establishes a new metallo hydrolase with β-lactamase activity which is structurally different from the conventional MBLs in P. difficile as a potential threat to clinical treatment.
Keywords: Pd metallo hydrolase, Peptoclostridium difficile, Metallo-beta-lactamases fold metallo hydrolase, Nosocomial infection, β-lactamase.
Detection of biofilm production among nosocomial isolates of multidrug-resistant Acinetobacter baumannii
Neha1 https://orcid.org/0000-0001-8791-6633 , Barnali Kakati2, Nupur Koul2, Vijay Kumar3
1Department of Microbiology, Himalayan School of Biosciences, Swami Rama Himalayan University, 2Department of Microbiology, Himalayan Institute of Medical Sciences, Swami Rama Himalayan University,
3Himalayan School of Biosciences, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Biofilm forming ability of multidrug-resistant pathogens contributes to the burden of antimicrobial resistance in health-care settings. Biofilm-producing carbapenem-resistant Acinetobacter baumannii (CRAB) have emerged as an opportunistic pathogen causing nosocomial infections limiting treatment options.
Aims and Objectives: This study was aimed at identifying biofilm-producing CRAB from various clinical specimens using phenotypic methods.
Material and Methods: This observational study was conducted in Department of Microbiology, Himalayan Institute Medical Sciences at Swami Rama Himalayan University, Dehradun. A total of consecutive 50 CRAB were included and subjected to phenotypic method for biofilm detection.
Results: All 50 isolates of A. baumannii were uniformly found resistant to carbapenems and identified as CRAB. Out of 50 CRAB isolates, 43 (94%) were identified as biofilm producers and 7 (6%) were identified as nonbiofilm producers by using microtiter plate method. Out of 43 biofilm-producing CRAB, 29 (67%) were isolated from intensive care unit and 14 (33%) isolated from wards.
Conclusion: High prevalence of biofilm producers was identified amongst CRAB isolates in our study. These findings will help us in identifying areas for implementation of biofilm eradication practices in critical settings reducing the burden of antimicrobial resistance by eradication of MDROs.
Keywords: Carbapenem-resistant Acinetobacter baumannii, Biofilm, Intensive care unit
Clinicoetiological and antimicrobial susceptibility profile of pediatric multi-drug-resistant infections
Shubh Kharbanda1, Nupur Koul2 https://orcid.org/0000-0002-3652-5584
1Department of Microbiology, Himalayan Institute of Medical Sciences,
2Department of Microbiology, Swami Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Background: Infections caused by multidrug-resistant organisms (MDRO) pathogens pose therapeutic challenges in pediatric and neonatal populations. Patients in Intensive care units (ICUs) are a major target population for hospital infectious pathogens due to several factors such as immune-compromised conditions, the use of broad-spectrum antibiotics, and the use of invasive devices. Alarming upsurge of infections caused by MDROs leading to high mortalities across all ages, especially newborns. Keeping the above in mind, this study was aimed at identifying the MDROs isolated from clinical specimens received from neonatal and pediatric ICUs, determining their antimicrobial susceptibility profile.
Methods: This observational study was conducted from September to November 2023. Identification and antimicrobial susceptibility testing of 100 multidrug-resistant pathogens recovered from various clinical specimens was done by automated VITEK-2 compact.
Results: The most common age group for pediatric infection was 29 days– 2 years (28%) with male preponderance (11:3). MDROs were most commonly recovered from cases of pediatric sepsis (33%) followed by lower respiratory tract infections (20%), renal infections (19%), and chronic suppurative otitis media (7%). The most common Gram-positive MDRO isolated was Staphylococcus species (22%). The most common Gram-negative MDRO isolated were Klebsiella pneumoniae (19%), Escherichia coli (13%), and Pseudomonas aeruginosa (12%). Out of 22 MDRO Staphylococcus species, all were found uniformly sensitive to nitrofurantoin (100%) and tigecycline (100%). MDRO Klebsiella spp. were found sensitive to fosfomycin (65%) and tigecycline (50%). MDRO E. coli were sensitive to fosfomycin (100%), tigecycline (85%), and cefoperazone-sulbactam (50%). MDRO P. aeruginosa were found sensitive to levofloxacin (50%) and ceftazidime (50%).
Conclusion: In our study, predominant pathogens causing MDRO infections in pediatric patients were Gram-negative organisms. Favorable antimicrobial susceptibility was found for fosfomycin, tigecycline, and levofloxacin. Findings from the study will be useful in appropriate antimicrobial selection for targeted therapeutic management of infections.
Keywords: Pediatric infections, multidrug-resistant organism, ESKAPE organisms
Clamping factor-mediated biofilm formation toward antimicrobial resistance of clinically isolated Staphylococcus aureus CHRFS5 from hemodialysis tunneled cuffed catheter tip
Rajsekhar Adhikary1 https://orcid.org/0000-0002-3074-1534
1Department of Biosciences and Bioengineering, Indian Institute of Technology, 2Centre for Nanotechnology, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Biofilm is defined as a multicellular bacterial community that grows on various surfaces or interfaces embedded within self-produced polymeric substances to induces virulence and facilitate adherence, colonization, immune evasion, and resistance to antimicrobial treatments. Biofilm formation mostly depends on the synthesis of an extracellular matrix that holds the constituent cells together. Surface colonized by Staphylococcus aureus with random biofilms forming ranging from different medical implants, such as catheters and orthopedic implants. One of the major factors of biofim formation is exopolysaccharide-mediated biofilm formation including polysaccharide intercellular adhesin or poly-N- acetylglucosamine, which play an important role in biofilm accumulation and maturation. Another major factor is clampling factor-mediated biofilm formatiob which controlled by two gene cassette, Clamping factor (Clf)A and ClfB. The present study focused on the biofilm formation pattern of clinical S. aureus CHRFS5 and also illustrate the deep insight on the evolutionary and structural aspects of the Clf cassette. The whole genome sequences of the clinical isolate possess distinct variations in its genetic make-up with the previous reports. The respective structural heterogeneity assessment among the Clf cassette considered both as a genetic tool for biofilm forming efficiency determination and virulance marker for clinical treatment and development of antimicrobial resistance.
Keywords: Antimicrobial resistance, Biofilm, Clamping factor, Hemodialysis tunneled cuffed catheter tip, Staphylococcus aureus CHRFS5
A review on bioremediation of emerging toxicants through microalgae
Vishal Rajput1 https://orcid.org/0000-0003-1182-6038 , Vivek Kumar1, Charu1
1Department of Himalayn School of Bio-sciences, Sri Rama Himalayan University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
During the last several decades, much emphasis has been paid to the control of environmental contamination produced by hazardous materials. Several approaches have been used to remove harmful components; however, these procedures have numerous drawbacks. The current endeavor focuses on an alternate biological agent that is plentiful in nature, namely microalgae. Microalgae may be found in both marine and freshwater ecosystems and have been shown to be critical to world ecology owing to their efficiency and taxonomically varied character. Microalgae are being investigated as a possible sink for hazardous compounds in the ecosystem. Microalgae have the potential to digest, collect, or absorb harmful substances found in the environment on a large scale. On the other side, the ever-increasing need for pesticides is causing major balance concerns in the soil and aquatic environments. Emerging toxicants now account for a large amount of pollution and are a severe threat to human health and the natural environment. Recent study has shown that bioremediation using microalgae is an innovative technique. Microalgal bioremediation has shown good possibilities as it is eco-friendly, effective, and inexpensive approach against toxicant buildup in ecosystem. In the present environment, bioremediation by microalgae aids biological remediation of developing toxicants in the ecosystem. The present manuscript focuses on culminate the stumbling block of bioremediation with the help of various microalgal strains and to emphasis on contemporary methodologies for effective bioremediation.
Keywords: bioremediation, emerging toxicants, microalgae, anthropogenic activities
Microbial warriors: Panchavalkala marvels’ stand against resistance - A deep dive
Geetika Pahuja1 https://orcid.org/0000-0002-4394-9203 , Meena Deogade2, Tanuja Nesari3
1Department of Dravyaguna, All India Institute of Ayurveda, New Delhi, India.
E-mail: nareshchettri15@gmail.com
Introduction: Anti-microbial resistance (AMR) represents a significant public health concern due to its high prevalence. Understanding the mechanisms underlying resistance is ground for developing effective therapeutic interventions. In parallel, many investigators have tried to reveal the potential antimicrobial properties of conventional medicines. Within this realm, Panchavalkala has emerged as a potential powerhouse, holding immense potential for the development of novel antibiotic. Panchavalkala, an Ayurvedic formulation which is composed of five plant barks, possesses a wide spectrum of bioactive compounds capable of combating pathogens in a manner distinct from conventional antibiotics. such as Panchavalkala, against Uropathogenic Escherichia coli (UPEC) strains.
Material and Methods: A comprehensive review of the published scientific literature indexed in electronic databases such as PubMed, Scopus, and Web of Science was conducted to gather evidence supporting these mechanisms.
Results: Panchavalkala, a traditional Ayurvedic formulation may exhibit potent antimicrobial activity due to its bioactive compounds. Tannins and flavonoids present in Panchavalkala may disrupt bacterial cell wall integrity, leading to structural alterations and compromising the cell wall integrity. Alkaloids and terpenoids may inhibit efflux pump activity, increasing intracellular antibiotic concentrations. Furthermore, flavonoids and alkaloids may interfere with essential metabolic pathways in UPEC, inhibiting nucleic acid synthesis and protein biosynthesis. Panchavalkala may also modulate virulence factors, reducing biofilm formation and adhesion.
Discussion: By targeting multiple pathways involved in developing resistance, Panchavalkala offers a promising approach to combat it.
Conclusion: Understanding the mechanisms of AMR and the mechanisms of action of Panchavalkala provides valuable insights for the development of novel therapeutic strategies against resistant infections.
Keywords: Antimicrobial resistance, Panchavalkala, Phytochemicals, Traditional medicine, Drug discovery, Reverse pharmacology
Assessing feasibility and utility of protocol-based antibiotic therapy in urological surgeries at a tertiary care center: A step toward antibiotic stewardship program
Vanya Singh1 https://orcid.org/0000-0002-5537-2499 , Omang2, Vikas Kumar Panwar3, Balram Ji Omar1, Ankur Mittal3
1Department of Microbiology, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, 2Department of Urology, MAX Saket, New Delhi,
3Department of Urology, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Introduction: Antimicrobial stewardship is essential to promote, improve, monitor, and evaluate the rational use of antimicrobials and preserve their future effectiveness. The purpose of this study was to assess the feasibility of implementation of protocol-based antibiotic therapy for Urological procedures to optimize rational antibiotic usage.
Objectives: (1) To study success rate of protocol-based antibiotic therapy for Urological procedures. (2) To identify underlying causal factors related to failure of protocol-based antibiotic therapy.
Material and Methods: A prospective observational study was done on patients undergoing urological surgeries at All India Institute of Medical Sciences Rishikesh from August 2020 to October 2021 with prior approval of the Institutional Ethics Committee. All the urological surgical cases were included except those in sepsis or undergoing relook surgery. Cases were classified as clean, clean contaminated, contaminated, or dirty. Protocol for decided based on American Urological Association guidelines. Data were analyzed using descriptive statistics. Any escalation or change in antibiotics or prolongation of duration of antibiotic therapy was considered as a failure of the protocol.
Results: A total of 145 patients were analyzed. Most of the surgeries were clean-contaminated (76.55%), followed by contaminated (11.72%) and clean (11.03%) cases. Prophylactic protocol was found to be successful in 82.07% (117/145) cases. Majority of the failed cases (16/26) had prolonged duration of surgery (>90 min).
Conclusion: Our study shows protocol-based surgical antibiotic prophylaxis is feasible and can prevent inappropriate use of antibiotics. Longer duration of antibiotics or higher antibiotics may be given for patients undergoing contaminated surgeries or those in sepsis/positive culture.
Keywords: Antimicrobial resistance, Antibiotic stewardship, Surgical antibiotic prophylaxis, Urological surgical procedures
Unveiling the antimicrobial potential of shatavari (Asparagus racemosus) leaves against uropathogens: A novel approach to urinary tract infection management
Mayuri Saini1, Shilpa Kukreti1, Neha Chauhan2 https://orcid.org/0000-0002-1687-0831
1Department of Microbiology, School of Basic and Applied Sciences, Shri Guru Ram Rai University, 2Department of Microbiology, School of Paramedical and Allied Health Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Urinary tract infections (UTIs) represent a prevalent global health concern, exacerbated by the emergence of multidrug-resistant (MDR) uropathogens, which pose significant challenges to conventional treatment modalities. In this context, exploring alternative therapeutic approaches becomes imperative. Shatavari (Asparagus racemosus), a traditional medicinal plant deeply rooted in Ayurvedic medicine, has garnered attention for its diverse health benefits. This study delves into the antimicrobial potential of Shatavari leaves against MDR uropathogens, offering a novel avenue for UTI management. Through a comprehensive review of literature, we examine the traditional use of Shatavari leaves and their bioactive constituents. Furthermore, we explore recent scientific evidence elucidating the antimicrobial activity of Shatavari leaf extracts against MDR uropathogens, including Escherichia coli, Klebsiella pneumoniae, and Enterococcus faecalis. Mechanistic insights into the antimicrobial action, including disruption of bacterial cell membranes and inhibition of virulence factors, are discussed. In addition, we highlight the potential synergistic effects of Shatavari leaf extracts with conventional antibiotics, addressing the pressing issue of antimicrobial resistance. Preclinical and clinical studies evaluating the efficacy and safety of Shatavari-based interventions in UTI management are reviewed. Moreover, safety considerations, future research directions, and challenges in translating these findings into clinical practice are addressed. Overall, this abstract underscores the promising antimicrobial activity of Shatavari leaves against MDR uropathogens, advocating for further research to validate its clinical utility in combating UTIs and antimicrobial resistance.
Keywords: Multidrug-resistant, Uropathogens, Shatavari, Virulence factors, Antimicrobial resistance
Unraveling Acinetobacter baumannii’s defense against oxidative stress: The role of superoxide dismutases in antibiotic resistance and host immunological response
Ashish Kumar Ray1 https://orcid.org/0009-0002-0179-5135 , Somok Bhowmik1, Snehalata Saini1, and Ranjana Pathania1
1Department of Biosciences and Bioengineering, Indian Institute of Technology, Roorkee, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
Bactericidal antibiotics affect the cellular redox state, promoting the generation of reactive oxygen species (ROS), which causes deoxyribonucleic acid damage and results in bacterial cell death. Thus ROS induction by antibiotic treatment contributes directly to antibiotic lethality. Bacteria utilize metalloenzymes such as superoxide dismutases (SODs) which break down endogenous and extracytoplasmic ROS. Bacterial SODs also help to mitigate from phagocytic cell-mediated oxidative stress and thus serve as a first line of defense in bacteria during infection. Due to the ability to cause infection in the immunocompromised patients, multidrug-resistant Acinetobacter baumannii has become a matter of concern in the health-care sector. Very little is known about the functional properties of A. baumannii SOD and its role in ROS-mediated secondary bactericidal antibiotic action and pathogenicity. To characterize SOD in A. baumannii, we purified SodB and SodC which are present in cytoplasm and periplasm of A. baumannii, respectively. We determined that SodB resides mostly in dimeric form, while SodC exists in both monomeric and dimeric forms. As previously stated, SODs are metalloenzymes that require a metal ion cofactor to function, and the cofactors used by SODs in A. baumannii are unknown. Next we wanted to determine which d-block transition metals are necessary for SodB and SodC and found that SodB and SodC utilize Mn2+ and Cu2+, respectively, for their functional activity. By delving into the multifaceted nature of SOD in A. baumannii, we aim to unravel its complexities in regulating ROS-mediated cellular damage and death.
Keywords: Reactive oxygen species, Acinetobacter baumannii, Superoxide dismutases, Antibiotics, Oxidative stress
Nanoengineered herbal gels: Revolutionizing shift in antimicrobial resistance wound therapies
Rahul Singh1,2, Satyendra Kumar Rajput1 https://orcid.org/0000-0002-0394-7369 , Gnana Ranjan2
1Department of Pharmaceutical Sciences, Gurukula Kangri Deemed to be University, Haridwar, 2Shri Guru Ram Rai University, Dehradun, Uttarakhand, India.
E-mail: nareshchettri15@gmail.com
The management of chronic wounds, especially in immunocompromised or diabetic populations, continues to pose significant challenges, necessitating evidence-based therapeutic interventions. Antimicrobials are used to prevent and treat infections in humans, animals, and plants. However, the antimicrobial resistance (AMR), including antibiotic resistance was identified as a grave problem during Sixty-eight World Health Assembly in May 2015. AMR occurs when microbes i.e., bacteria, viruses, fungi, and parasites no longer respond to the antimicrobials, i.e., antibiotics, antivirals, antifungals, and anti-parasitic drugs, thus antibiotics and other antimicrobial drugs become ineffective; and infections become increasingly difficult or impossible to treat. Moreover, herbal remedies, deeply rooted in centuries-old traditions, tap into the therapeutic properties of plants, offering a culturally accepted and trusted alternative. Recent investigations highlight the capacity of herbal extracts to augment wound healing processes, leveraging mechanisms such as antioxidant activity, promotion of angiogenesis, and stimulation of collagen synthesis. Integration with nanotechnology further enhances the delivery and efficacy of herbal medications, surmounting obstacles such as poor solubility. This paper adopts a structured approach, defining a focused research question and establishing clear inclusion and exclusion criteria. The convergence of nanotechnology and herbal medicine heralds a paradigm shift in wound care, ushering in a new era characterized by heightened precision, sustained efficacy, and minimized side effects. The integration of herbal nano gels holds immense promise for optimizing wound treatment outcomes, surmounting the limitations of conventional methods, and contributing to the ongoing evolution of wound care strategies. In conclusion, herbal nano gels are pioneering modalities in wound healing.
Keywords: Antimicrobial resistance, Medicinal plants, Advanced wound therapies, Nano drug delivery, Bioactive compounds, Wound care innovation




