Translate this page into:
Detection of carbapenemase-producing Pseudomonas aeruginosa by phenotypic and genotypic methods in a tertiary care hospital of East India
Address for correspondence: Dr. Baijayantimala Mishra, Department of Microbiology, All India Institute of Medical Sciences, Bhubaneswar - 751 019, Odisha, India. E-mail: bm_mishra@hotmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Carbapenemase-producing Pseudomonas aeruginosa is a serious threat in hospital infection due to its multidrug resistance.
AIM:
The aim of the study was to determine the frequency of carbapenem resistance in clinical isolates of Pseudomonas aeruginosa and detect the presence of carbapenemase enzymes in carbapenem-resistant P. aeruginosa (CRPA) isolates by phenotypic and genotypic methods.
MATERIAL AND METHODS:
Double-disk synergy test [DDST] and combined disk synergy test [CDST]) was performed in CRPA isolates and the prevalence of blaKPC, blaNDM-1, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM, and blaOXA-48 was determined.
RESULTS:
Of 559 isolates included in the study, a total of 102 isolates were resistant to carbapenem that accounted for overall 18.24% (102/559) prevalence. Of these 102 isolates, 89 (87.25%) isolates were positive by DDST and 95 (93.17%) isolates were positive by CDST. Of 102 CRPA isolates, blaVIM was detected in 30 isolates (30/102, 29.1%), followed by blaNDM-1 in 29 (29/102, 28.4%) isolates and blaSIM and blaGIM in 6 isolates each (6/102, 5.8%). A combination of two carbapenemase genes was detected in 12 isolates, with six (6/102, 5.88%) CRPA isolates harboring with both blaVIM and blaNDM-1 genes. Four isolates were found to harbor a combination of three carbapenem-resistant genes.
CONCLUSION:
A high rate of carbapenemase production was observed in P. aeruginosa. Coproducers of multiple carbapenemases are also a cause of concern. An in-depth understanding of molecular mechanisms of resistance will be helpful in optimizing patient management and hospital infection control.
Keywords
Carbapenemases
genotypic methods
phenotypic methods
Pseudomonas aeruginosa
Introduction
Carbapenem-resistant Pseudomonas aeruginosa (CRPA) is listed as an organism posing a serious threat by the Centers for Disease Control and Prevention.[1] In India, up to 40% P. aeruginosa isolates are shown to be carbapenem resistant.[2] Carbapenem resistance in P. aeruginosa has shown to be multifactorial, including production of carbapenemase, overexpression/overproduction of efflux pump, and porin loss. Carbapenemases in P. aeruginosa belong to three molecular classes, for example, class A (blaKPC), class B (blaIMP, blaVIM, blaNDM, blaSIM, blaGIM, and blaSPM), and class D (blaOXA-48) genes. The molecular mechanism of carbapenem resistance in P. aeruginosa has been studied in very few studies restricted to North and South India.[23]
The aim of the present study was to determine the production of carbapenemase enzymes in CRPA isolates by phenotypic methods (double-disk synergy test [DDST] and combined disk synergy test [CDST]) and to determine the prevalence of blaKPC, blaNDM-1, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM, and blaOXA-48. The susceptibility profile of P. aeruginosa to other antipseudomonal antibiotics was also investigated.
Materials and Methods
This study was conducted at the microbiology department of a tertiary care teaching hospital from January 2017 to May 2018. All consecutive nonduplicate isolates of P. aeruginosa from different clinical samples (blood, respiratory sample, pus, urine, sterile body fluids, and other samples) resistant to meropenem and/or imipenem by disk diffusion method were included in the study. The study was conducted after ethical clearance from the Institutional Ethics Committee (ref no-IEC/AIIMS BBSR/PG Thesis/2017–18/3).
The initial isolation and identification of P. aeruginosa were carried out by standard bacteriological procedures. The following antimicrobials were tested by disk diffusion: ceftazidime (30 μg), cefepime (30 μg), piperacillin–tazobactam (100 μg/10 μg), ticarcillin–clavulanate (75 μg/10 μg), imipenem (10 μg), meropenem (10 μg), doripenem (10 μg), amikacin (30 μg), gentamicin (10 μg), netilmicin (30 μg), tobramycin (10 μg), ciprofloxacin (5 μg), and levofloxacin (5 μg) (HiMedia Laboratories Pvt. Ltd., Mumbai, Maharashtra, India). The antimicrobial susceptibility was interpreted as per the Clinical and Laboratory Standards Institute 2017 guidelines using P. aeruginosa ATCC strain 27853 as quality control strain.[4] In addition, imipenem and meropenem minimum inhibitory concentration was determined by agar dilution and E-test (HiMedia Laboratories Pvt. Ltd., Mumbai, Maharashtra, India).
Isolates with imipenem/meropenem/doripenem MIC ≥8 μg/ml by agar dilution and E-test were screened for carbapenemase production by DDST and CDST by the following methods.
-
Double-disk synergy test:[5] As described by Lee et al., this test was carried out using ethylenediaminetetraacetic acid (EDTA) disk (10 μl of 0.5 M EDTA) and the imipenem disk (10 μg) using the standard method.[5] The production of large synergistic inhibitory zones was interpreted as test positive for metallo-beta-lactamase (MBL)
-
Combined disk synergy test:[6] As described by Yong et al., this test was carried out using two imipenem commercial disks with and without EDTA (10 μl of 0.5 M EDTA).[6] The zone of inhibition diameter with imipenem + EDTA disk ≥7 mm than the imipenem disk alone was considered positive for MBL production.
Molecular detection of carbapenemase gene
Multiplex polymerase chain reaction (PCR) for the presence of blaKPC, blaNDM-1, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM, and blaOXA-48 was carried out according to the method published by Ellington et al.[7] and Kumarasamy et al.[8] Briefly, DNA extraction from P. aeruginosa isolates was carried out using QIAamp DNA Mini kit spin-column method (QIAGEN, Hilden, Germany) as per manufacturer's instruction. Two sets of multiplex PCR were done using KAPA2G Fast Multiplex PCR Kit (2X) as per manufacturer's instructions (Kapa Biosystems, Cape Town, South Africa). The first set of multiplex PCR detected blaIMP, VIM, GIM, SPM, and SIM and the second set of multiplex PCR detected blaNDM-1, OXA-48, and KPC. The PCR reactions consisted of a final reaction volume of 25 μl containing 5 μl of template DNA, 12.5 μl of 2X KAPA2G Fast Multiplex Master Mix, and 0.5 μl of each oligonucleotide primer. Amplification was done with initial denaturation step at 95°C for 3 min, followed by denaturation at 95°C for 15 s, annealing at 60°C for 30 s, extension at 72°C for 30 s, and a final extension step at 72°C for 7 min. The product was visualized by 1.5% agarose gel electrophoresis, stained with ethidium bromide, and was viewed under automated gel documentation system (Syngene G: Box, Syngene, Cambridge, U.K.) using ultraviolet illumination. The primer sequences and the product sizes[78] are given in Table 1.
| Primer | Primer sequence (5’-3’) | Product size (bp) |
|---|---|---|
| IMP family-F | GGA ATA GAG TGG CTT AAY TCT C | 188 |
| IMP family-R | CCA AAC YAC TAS GTT ATC T | |
| VIM family-F | GAT GGT GTT TGG TCG CAT A | 390 |
| VIM family-R | CGA ATG CGC AGC ACC AG | |
| GIM-1-F | TCG ACA CAC CTT GGT CTG AA | 477 |
| GIM-1-R | AAC TTC CAA CTT TGC CAT GC | |
| SPM-1A-F | AAA ATC TGG GTA CGC AAA CG | 271 |
| SPM-1A-R | ACA TTA TCC GCT GGA ACA GG | |
| SIM-1-F | TAC AAG GGA TTC GGC ATC G | 571 |
| SIM-1-R | TAA TGG CCT GTT CCC ATG TG | |
| NDM-1 F | ACC GCC TGG ACC GAT GAC CA | 264 |
| NDM-1 R | GCC AAA GTT GGG CGC GGT TG | |
| OXA-48-F | TTGGTGGCATCGATTATCGG | 744 |
| OXA-48-R | GAGCACTTCTTTTGTGATGGC | |
| KPC-F | ATGTCACTGTATCGCCGTCT | 893 |
| KPC-R | TTTTCAGAGCCTTACTGCCC |
Statistical analysis
The sensitivity and specificity of CDST and DDST were calculated by forming 2 × 2 contingency tables and genotypic tests being considered as a gold standard.
Results
A total of 559 consecutive nonduplicate isolates of P. aeruginosa were recovered from various clinical samples received in the department of microbiology during the study period (January 2017–May 2018). Of 559 isolates of P. aeruginosa, a total of 102 isolates (102/559, 18.2%) were resistant to carbapenem (MIC ≥8 μg/ml). Of these 102 isolates, 19 (19/102, 18.6%) were from various intensive care units, 24 (24/102, 23.5%) were from various outpatient departments, and rest 59 (59/102, 57.8%) were from various admitted patients. CRPA was isolated from pus (38/102, 37.2%), followed by urine (31/102, 30.3%), respiratory samples (25/102, 24.5%), blood (3/102, 2.9%), and other samples (5/102, 4.9%), respectively. Among the various antipseudomonal antibiotics tested in CRPA isolates, the maximum susceptibility was observed for piperacillin–tazobactam (17/102, 16.6%), followed by ceftazidime (12/102, 11.7%) and aminoglycosides (8/102, 7.8%). Ticarcillin–clavulanate with a percentage susceptibility of 2.9% (3/102) was found to be least susceptible. All CRPA isolates were found to be resistant to ciprofloxacin and levofloxacin. Of 102 CRPA isolates, CDST and DDST were positive in 95 (93.1%) and 89 (87.2%), respectively, as shown in Table 2. Figures 1 and 2 depict the positive CDST and DDST, respectively.
| Type of carbapenemase gene identified | Number of isolates (%), (n=102) | CDST+ | CDST− | DDST+ | DDST− |
|---|---|---|---|---|---|
| blaNDM-1 | 19 (18.62) | 19 | 0 | 15 | 4 |
| blaVIM | 17 (16.62) | 16 | 1 | 14 | 3 |
| blaSIM | 6 (5.88) | 5 | 1 | 6 | 0 |
| blaGIM | 6 (5.88) | 4 | 2 | 5 | 1 |
| blaVIM+blaNDM-1 | 6 (5.88) | 6 | 0 | 5 | 1 |
| blaVIM+blaGIM | 3 (2.94) | 3 | 0 | 3 | 0 |
| blaGIM+blaNDM-1 | 2 (1.96) | 2 | 0 | 2 | 0 |
| blaGIM+blaSPM | 1 (0.98) | 1 | 0 | 0 | 1 |
| blaVIM+blaGIM+blaSIM | 1 (0.98) | 0 | 1 | 1 | 0 |
| blaVIM+blaSPM+blaNDM-1 | 1 (0.98) | 1 | 0 | 0 | 1 |
| blaVIM+blaGIM+blaNDM-1 | 1 (0.98) | 1 | 0 | 1 | 0 |
| blaVIM+blaGIM+blaSPM | 1 (0.98) | 1 | 0 | 1 | 0 |
| Total | 64 | 59 | 5 | 53 | 11 |
CDST=Combined disk synergy test
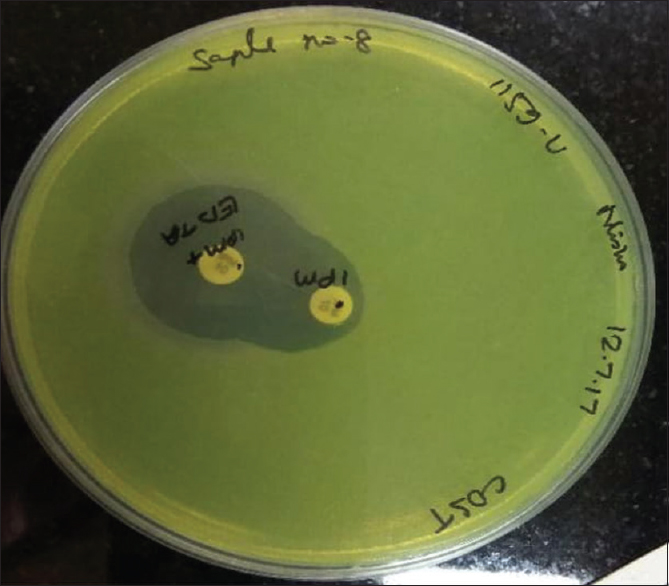
- Isolate showing positive carbapenemase-producing Pseudomonas aeruginosa test with ≥ 7 mm increase in the zone of inhibition of imipenem + ethylenediaminetetraacetic acid as compared to imipenem alone
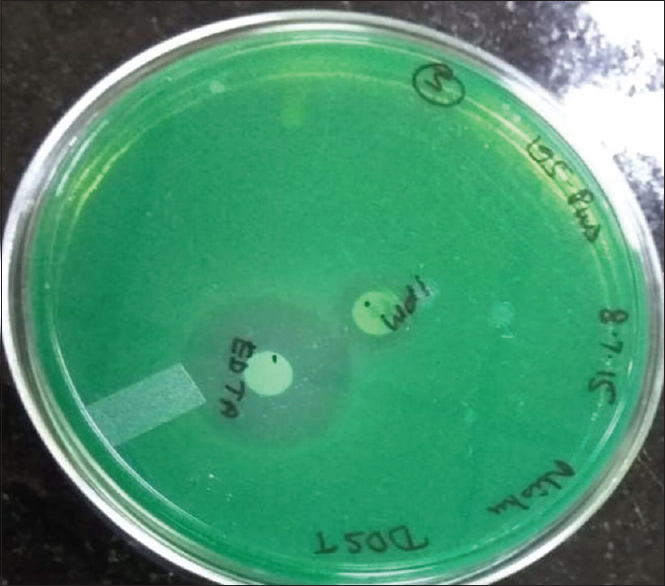
- Isolate showing positive double-disk synergy test with enhanced zone of inhibition around ethylenediaminetetraacetic acid as compared to imipenem
Of 102 CRPA isolates, blaVIM was detected in 30 isolates (30/102, 29.1%), followed by blaNDM-1 in 29 (29/102, 28.4%) isolates and blaSIM and blaGIM in 6 isolates each (6/102, 5.8%). A combination of two carbapenemase genes were detected in 12 isolates, with six (6/102, 5.88%) CRPA isolates harboring with both blaVIM and blaNDM-1 genes. Four isolates were found to harbor a combination of three carbapenem-resistant genes [Figure 3]. No CRPA isolates were found to possess blaIMP, blaKPC, and blaOXA-48. The distribution of various carbapenemase genes in 102 CRPA isolates is provided in Table 2. Five isolates of CRPA that were positive for NDM-1 were sent for sequencing by Sanger's method to Eurofins Genomics India Pvt. Ltd., Bengaluru, Karnataka, India, and interpreted using BLAST software (NIH, USA), of which three were NDM-1 and two were found to be NDM-5. The sensitivity of DDST and CDST was found calculated taking the presence of any MBL gene as a reference test and was found to be 81.35% and 92.18%, respectively, as shown in Table 3.
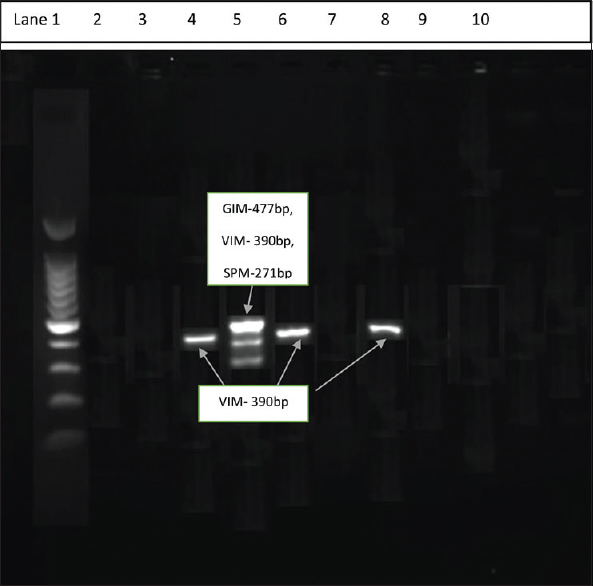
- Multiplex polymerase chain reaction - Lane 1 showing 100 bp ladder; Lanes 4, 6, and 8 showing blaVIM-390 bp; and Lane 5 showing blaGIM + blaVIM + blaSPM
| Type of test | Positive (%) | Negative (%) | Sensitivity (%) |
|---|---|---|---|
| DDST test | 89 (87.25) | 13 (12.75) | 81.35 |
| CDST test | 95 (93.17) | 7 (6.83) | 92.18 |
DDST=Double-disk synergy test, CDST=Combined disk synergy test
Discussion
Carbapenem resistance was noted in 18.2% P. aeruginosa isolates in our study. In the Indian Council of Medical Research scoping document, the prevalence of CRPA isolates across Indian hospitals is shown to be 42%–47% in the years 2014 and 2015.[9] In a recent single-center study from North India, the prevalence of CRPA is shown to be 72.5% in admitted patients.[10] The relatively lower prevalence of CRPA in our study could be due to the higher proportion of isolates being included from outpatient settings.
In our study, the sensitivity of DDST and CDST was found to be 81.35% and 92.18%, respectively (taking the presence of any MBL gene as a reference test). The sensitivity of CDST for MBL detection was shown to be 86.7% and specificity of 51.1% in a previous study by Peter et al. in 2014.[11] The higher sensitivity of CDST compared to DDST, as observed in our study, is also described previously.[12] In the present study, the prevalence of MBL was 15.74% by phenotypic method (CDST), whereas the prevalence by genotypic method for MBL was found as 10.19%. The concordance rate between CDST and genotypic method of MBL detection in our study was found to be 92.18%. The overall rate of concordance between phenotypic and genotypic tests for the detection of MBL was 98% in a study by Kazi et al.[13]
In our study, 62.74% CRPA isolates had at least one carbapenemase gene. blaVIM was the most prevalent gene in CRPA (30/64, 46.87%), followed by blaNDM-1 (29/64, 45.31%). blaVIM and blaNDM are shown to the most prevalent carbapenemase gene in the majority of Indian studies.[231214] In the study by Ellapan et al. in 2018, blaVIM and blaNDM were present in 23.1% and 17.3% of 156 CRPA isolates.[15] In another study by Mohanam and Menon in 2017, blaVIM and blaNDM were present in 32% and 27% of 213 CRPA isolates.[16] In a multicentric study from India, blaVIM and blaNDM were the most frequent carbapenemases in CRPA isolates with regional differences in the carbapenemase profile across the study sites.[2] The incidence of CRPA has increased worldwide, and they pose a potential risk for therapeutic failure with the empirical treatments currently in place. In this study, no blaKPC, blaIMP, and blaOXA-48 were identified. blaKPC and blaOXA-48 were also absent in 156 CRPA isolates analyzed by Ellapan et al.[15] In another multicentric study by Khurana et al., blaKPC and blaOXA-1 were present in 43% and 56% of CRPA isolates from North India but were absent in CRPA isolates collected from South India.[17] In the present study, 15 CRPA isolates had coexistence of two or three carbapenem-resistant genes. blaVIM + blaNDM-1 was the most common combination (6/102, 5.88%), followed by blaVIM + blaGIM (3/102, 2.94%). The coproduction of blaVIM + blaNDM-1 had previously been described in 7.1% CRPA isolates by Ellapan et al.[15] The coexistence of blaVIM + blaNDM-1 and blaKPC-2 and blaNDM-1 in CRPA isolates from India is also described previously by Paul et al.[1819]
No carbapenem-resistant gene was detected in 38 (38/102, 37.25%) CRPA isolates in our study. Other carbapenem mechanisms such as efflux pump and porin loss could have contributed for carbapenemase resistance, which were not studied in the present study. Loss of OprD porin and overexpression of mexA gene is also found in studies from India either as a standalone mechanism or in association with the presence of blaVIM.[15]
In our study, fluoroquinolones did not have any activity against CRPA isolates. Polymyxin B and colistin were found to be susceptible in all isolates of P. aeruginosa. There are recent studies of colistin resistance in <5% P. aeruginosa isolates from India, which warrant judicious use of this last-resort antibiotic.[14] Among beta-lactam and beta-lactamase inhibitor combination, the maximum susceptibility was observed for piperacillin–tazobactam (16.67%), followed by ceftazidime with 11.76% and 2.94% for ticarcillin–clavulanate which was found to be least susceptible. Carbapenem-resistant and cephalosporin-susceptible P. aeruginosa is a rare phenotype. Previous carbapenem therapy and decreased OprD expression and efflux system overexpression are cited as possible mechanisms of this phenotype. The clinical utility in this group is, however, debatable.[20]
Conclusion
A high rate of carbapenemase production was observed in P. aeruginosa. Coproducers of multiple carbapenemases are also a cause of concern. An in-depth understanding of molecular mechanisms of resistance will be helpful in optimizing patient management and hospital infection control.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Centers for Disease Control and Prevention, Office of Infectious Diseases. Antibiotic Resistance Threats in the United States, 2013. 2013. Atlanta, GA: Centers for Disease Control and Prevention, Office of Infectious Diseases; Available from: http://www.cdc.gov/drugresistance/threat-report-2013
- [Google Scholar]
- Dominance of international high-risk clones in carbapenemase-producing Pseudomonas aeruginosa: Multicentric molecular epidemiology report from India. Indian J Med Microbiol. 2018;36:344-51.
- [Google Scholar]
- Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J Med Microbiol. 2016;34:433-41.
- [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI Document M100 (27th ed). Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- [Google Scholar]
- Evaluation of the hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41:4623-9.
- [Google Scholar]
- Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40:3798-801.
- [Google Scholar]
- Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59:321-2.
- [Google Scholar]
- Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597-602.
- [Google Scholar]
- Scoping report on antimicrobial resistance in India. Washington, DC: Center for Disease Dynamics, Economics and Policy; 2017.
- [Google Scholar]
- Phenotypic genotypic profile of antimicrobial resistance in Pseudomonas species in hospitalized patients. Indian J Med Res. 2019;149:216-21.
- [Google Scholar]
- Evaluation of phenotypic detection methods for metallo-β-lactamases (MBLs) in clinical isolates of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2014;33:1133-41.
- [Google Scholar]
- An evaluation of four different phenotypic techniques for detection of metallo-beta-lactamase producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26:233-7.
- [Google Scholar]
- Dual-tubed multiplex-PCR for molecular characterization of carbapenemases isolated among Acinetobacter spp. and Pseudomonas spp. J Appl Microbiol. 2015;118:1096-102.
- [Google Scholar]
- Newer ß-Lactam/ß-Lactamase inhibitor for multidrug-resistant gram-negative infections: Challenges, implications and surveillance strategy for India. Indian J Med Microbiol. 2018;36:334-43.
- [Google Scholar]
- Coexistence of multidrug resistance mechanisms and virulence genes in carbapenem-resistant Pseudomonas aeruginosa strains from a tertiary care hospital in South India. J Glob Antimicrob Resist. 2018;12:37-43.
- [Google Scholar]
- Coexistence of metallo-beta-lactamaseencoding genes in Pseudomonas aeruginosa. Indian J Med Res. 2017;146:S46-52.
- [Google Scholar]
- Molecular epidemiology of beta-lactamase producing nosocomial Gram-negative pathogens from North and South Indian hospitals. J Med Microbiol. 2017;66:999-1004.
- [Google Scholar]
- Co-Carriage of blaKPC-2 and blaNDM-1 in clinical isolates of Pseudomonas aeruginosa Associated with hospital infections from India. PLoS One. 2015;10:e0145823.
- [Google Scholar]
- Occurrence of co-existing bla VIM-2 and blaNDM-1 in clinical isolates of Pseudomonas aeruginosa from India. Ann Clin Microbiol Antimicrob. 2016;15:31.
- [Google Scholar]
- Carbapenem-resistant and cephalosporin-susceptible Pseudomonas aeruginosa: A notable phenotype in patients with bacteremia. Infect Drug Resist. 2018;11:1225-35.
- [Google Scholar]





