Translate this page into:
Detection of hypervirulent Klebsiella pneumoniae among clinical isolates from select infections using virulence gene markers
*Corresponding author: Sujatha Sistla, Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India. sujathasistla@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Rajamanickam S, Manoharan M, Bhaskar M, Sistla S. Detection of hypervirulent Klebsiella pneumoniae among clinical isolates from select infections using virulence gene markers. J Lab Physicians. doi: 10.25259/JLP_241_2024
Abstract
Objectives
Hypervirulent Klebsiella pneumoniae (hvKp) is a distinct pathotype of K. pneumoniae associated with community-acquired infections, and it generally exhibits susceptibility to antimicrobial agents. The objective of the study was to identify hvKp and determine the antibiotic susceptibility profiles of these isolates.
Materials and Methods
A total of 150 K. pneumoniae isolates were obtained from community-acquired infections. All isolates were subjected to string test and antimicrobial susceptibility test using Kirby–Bauer disc diffusion. Final confirmation was done using polymerase chain reaction (PCR), targeting three hvKp-specific virulence genes (p rmpA, iucA, and peg344).
Statistical analysis
The associations between the string test and virulence genes were assessed using the Chi-square test. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the string test were calculated using PCR as the gold standard.
Results
Among the 150 isolates, 62 (41.3%) were identified as hvKp. Abscesses (29) and pneumonia (22) were the most common clinical conditions, with neck abscesses as the predominant site. Of the 62 hvKp isolates, only 44 were string test-positive. Surprisingly, 22 string test-positive isolates lacked these genes, while 18 string test-negative isolates carried virulence genes. The sensitivity, specificity, PPV, and NPV of the string test were 66.7%, 78.5%, 71%, and 75%, respectively. Three hvKp isolates were multidrug-resistant.
Conclusions
Accurate detection of hvKp is critical in clinical settings due to its association with severe community-acquired infections, including deep-seated abscesses and pneumonia. The string test lacks both sensitivity and specificity, making it unreliable for hvKp identification. Therefore, the detection of virulence genes remains the gold standard. Early and precise diagnosis is essential for guiding appropriate treatment, including targeted antibiotic therapy and necessary drainage procedures for abscesses. The presence of multidrug resistance in certain isolates is a cause for concern, necessitating strict infection control measures to prevent the spread of such strains.
Keywords
Hypermucoviscous
Hypervirulent Klebsiella pneumoniae
Multidrug-resistant hypervirulent Klebsiella pneumoniae
String test
Virulence genes
INTRODUCTION
Klebsiella pneumoniae is a common cause of both nosocomial and community-acquired infections, including pneumonia, bacteremia, urinary tract infections, and pyogenic infections.[1,2] This pathogen constantly evolves due to its ability to acquire new genes leading to the emergence of two distinct pathotypes, namely, classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp).[1] The majority of K. pneumoniae infections are attributed to classical (cKP) strains, which primarily affect chronically ill patients residing in hospitals and long-term care facilities. These strains have shown increasing resistance to antibiotics, including carbapenems, leading to challenging-to-treat infections.[3]
In contrast, hvKp infections have been acquired within the community. It is an evolving pathomorphotype, also known as Hypermucoid or Hypermucoviscous Klebsiella, that infects healthy individuals of all age groups and causes infected patients to develop infections at multiple sites and metastatic spread of infection.[1,2] hvKp was first reported in 1986 in Taiwan from a case of pyogenic liver abscess (PLA) associated with endophthalmitis.[4] However, it has now emerged as a pathogen capable of causing deep-seated visceral abscesses (such as in the liver and spleen), necrotizing fasciitis, meningitis, endophthalmitis, community-acquired pneumonia, and even hospital-acquired infections with a multidrug-resistant phenotype.[5]
hvKp has been associated with several important characteristics. One is the excessive production of exopolysaccharides, resulting in a hypermucoviscous phenotype.[6] The string test, based on hypermucoviscous phenotype, is widely used as a screening test for the detection of hvKp strains, as most of them produce hypermucoviscous colonies.[7] In a study from India, Raj et al. stated that the sensitivity of the string test varies, ranging from 51% to 98%.[2] Another characteristic is the presence of large virulence plasmids (pk2044 and pLVPK) and integrative conjugative elements carrying specific genes, such as iucA (aerobactin siderophore biosynthesis), peg344 (putative transporter), and rmpA (regulator of mucoid phenotype).[1]
These hvKp strains are more commonly susceptible to many antibiotics, including carbapenems and cephalosporins. However, recently, emerging multidrug-resistant hvKp strains have been reported.[7,8] At present, there is a lack of awareness regarding this emerging pathotype among both clinicians and microbiologists, which has resulted in its under-recognition and reporting. In addition, there is a paucity of data on the prevalence, type of infections, and the presence of multidrug resistance (MDR) among hvKp isolates from India. Therefore, the present study was undertaken to identify hvKp and determine the antibiotic susceptibility profiles of these isolates.
MATERIALS AND METHODS
The study was approved by the Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER) Institute Human Ethics Committee (IEC; JIP/IEC/2019/332). A total of 150 K. pneumoniae isolates were collected from various clinical samples, such as pus, respiratory samples, sterile fluids, and blood, during the study period of January 2020–June 2021 in the Department of Microbiology, JIPMER.
Isolates were selected based on the inclusion criteria, which required consecutive, non-duplicate clinical isolates of K. pneumoniae from select infections, such as community-acquired pneumonia, other respiratory infections, bacteremia, deep-seated or multiple abscesses, necrotizing fasciitis, meningitis, and endophthalmitis. All suspected strains were identified based on the clinical diagnosis recorded in the request form and samples were collected within 48 hours of admission. Isolates from confirmed hospital-acquired infections were excluded from the study.
Isolates were identified using Matrix-Assisted Laser Desorption/Ionization-Time of Flight mass spectrometry (VITEK® MS, BioMerieux, France) based on the manufacturer’s instructions. All the isolates were subjected to phenotypic and genotypic characterization methods to detect hvKp.
Phenotypic method
String test
The string test, which involves the formation of a viscous string measuring >5 mm was performed on mucoid colonies present on agar plates.[9]
Antimicrobial susceptibility testing
All the isolates were tested for susceptibility to amikacin (30 µg), ceftazidime (30 µg), ceftriaxone (30 µg), ciprofloxacin (5 µg), cefoperazone+sulbactam (75/10 µg), meropenem (10 µg), levofloxacin (5 µg), and piperacillin/tazobactam (100/10 µg) using the Kirby–Bauer disc diffusion method. American Type culture collection (ATCC) Escherichia coli 25922 and ATCC Pseudomonas aeruginosa 27853 were used as quality control strains for all the tested antibiotics and the results were interpreted according to the Clinical and Laboratory Standards Institute 2020 guidelines.[10]
Detection of hypervirulence genes using polymerase chain reaction (PCR)
DNA was extracted from isolates using a Mericon DNA kit (Qiagen, Germany) as per the manufacturer’s instructions. Conventional Uniplex and Duplex PCR were performed in all the isolates to detect the presence of three hypervirulence genes in the plasmid, namely, peg344 , iucA, and rmpA. The presence of any one of the tested genes was considered indicative of hvKp. The details of primer sequences and cycling conditions with amplicon base pair size are given in Table 1.
| Genes | Primer sequence (5’-3’) | Amplicon size (bp) | PCR Condition | Reference |
|---|---|---|---|---|
| iucA | F: AATCAATGGCTATTCCCGCTG R: CGCTTCACTTCTTTCACTGACAGG |
239 | 95°C for 2 min; 25 cycles of 95°C for 30 s; 54°C for 30 s; 72°C for 1 min and final elongation at 72°C for 5 min | [5] |
| peg344 | F: CTTGAAACTATCCTCCAGTC R: CCAGCGAAAGAATAACCCC |
508 | ||
| rmpA | F: GAGTAGTTAATAAATCAATAGCAAT R: CAGTAGGCATTGCAGCA |
332 |
PCR: Polymerase chain reaction, bp: base pair
Statistical analysis
Statistical analysis was performed using STATA version 11.0 (Stata Corp LP, TX, USA). Associations between the string test and virulence genes were assessed using Pearson Chi-square test. P ≤ 0.05 was considered as statistically significant. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of string test were calculated using PCR as the gold standard.
RESULTS
Among the 150 K. pneumoniae isolates, 62 (41.3%) were identified as the hvKp pathotype, while 88 (58.7%) were classified as the classical pathotype based on the presence of virulence genes. Out of these 62 confirmed hvKp isolates, only 44 were string test positive, while 18 isolates were negative for the string test. However, there were 22 string test-positive isolates that were negative for virulence genes (not hvKp).
Distribution of virulence genes
Among the 62 confirmed hvKp isolates, 55 (88.7%) carried all three genes, while the iucA gene alone was detected in only four isolates (6.5%). Two isolates (3.2%) carried the rmpA gene and only one (1.6%) harbored the peg344 gene. Among the 18 hvKp isolates that tested negative in the string test, the rmpA gene was found in 17 isolates, while only one isolate lacked this gene. Interestingly, only one isolate of hvKp negative for this gene exhibited a positive string test. The presence of any one of these three genes indicated hvKp [Figures 1 and 2]. Three randomly selected hvKp isolates were confirmed for the presence of the three genes (iucA, rmpA, and peg344) using Sanger sequencing. The nucleotide sequences were compared to previously published sequences from the National Center for Biotechnology Information (NCBI) database and were found to be similar. The accession numbers are as follows; iucA – CP091059, peg344 – CP054781, and rmpA – MT330316, respectively.
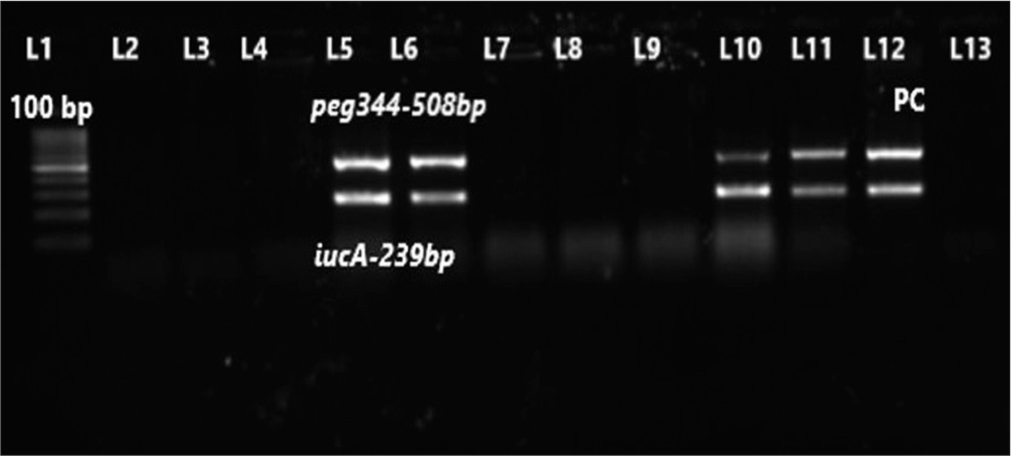
- Gel picture of duplex polymerase chain reaction for iucA and peg344 genes. Lane-1 (L1) – 100 bp molecular marker; L2-L4 and L7-L9 – test isolates negative for iucA and peg344 genes; L5, L6, L10, and L11 – test isolates positive for both iucA (239 bp) and peg344 genes (508 bp), respectively; L12-PC for iucA and peg344 genes; L13-Negative control.
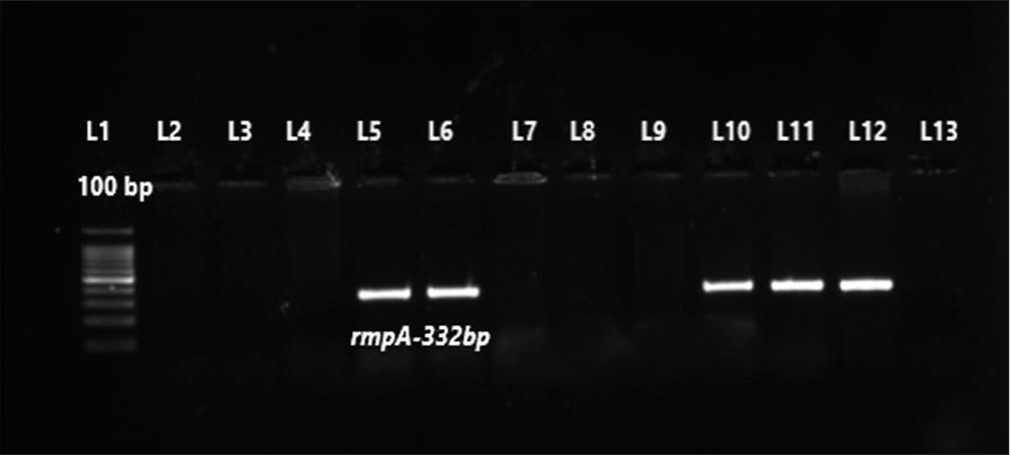
- Gel picture of uniplex polymerase chain reaction for rmpA gene. Lane-1 (L1) – 100 bp molecular marker; L2-L4 and L7- L9 – test isolates negative for rmpA; L5, L6, L10 and L11 – test isolates were positive for rmpA gene (332 bp); L12-Positive control for rmpA gene (332 bp); L13-Negative control.
Validity of string test for identification of hvKp
The string test exhibited moderate sensitivity (66.7%) and a PPV (71%) as well as slightly higher specificity (78.5%) and an NPV of 75% when compared to PCR for virulence genes. The string test showed a weak kappa agreement of 0.45 with a confidence interval of 0.29–0.61.
Demographic and clinical profile of patients with hvKp infection
The mean age of patients with hvKp infection was 48.03 ± 2.13 years. Among hvKp infections, abscesses were the most common presentation (29/62, 51.8%), followed by pneumonia (22/61, 36.1%) and perforation peritonitis (7/17, 41.2%). Deep-seated abscesses included liver, perinephric, splenic, pancreatic, and lung abscesses. Among the abscesses, neck abscess (6/56; 10.7%) was the most common presentation in hvKp infection. There was a strong association between hvKp infection and abscesses (P = 0.04). On the other hand, cKp had a significant association with diabetic foot infections (P = 0.03) [Table 2].
| Serial No | Clinical condition | hvKp number (%) | cKp number (%) | P-value |
|---|---|---|---|---|
| 1 | Pneumonia (61) | 22 (36) | 39 (64) | 0.285 |
| 2 | Abscesses (56) | 29 (51.8) | 27 (48.2) | 0.04 |
| 3 | Perforation peritonitis (17) | 7 (41.2) | 10 (58.8) | 0.998 |
| 4 | Bronchiectasis (3) | 3 (100) | - | 0.067 |
| 5 | Diabetic foot (10) | 1 (10) | 9 (90) | 0.03 |
| 6 | Bacteremia (2) | - | 2 (100) | 0.34 |
| 7 | Necrotizing fasciitis (1) | - | 1 (100) | 0.59 |
| Monomicrobial infection (93) | 50 (53.8) | 43 (46.2) | <0.001 | |
The numbers in parentheses under “Clinical condition” represent the total number of cases for each respective condition. P-values were calculated using the chi-square test or Fisher’s exact test, as appropriate. P≤ 0.05 is considered as statistically significant and is highlighted in bold. hvKp: Hypervirulent Klebsiella pneumoniae, cKp: Classical K. pneumoniae.
Among the 62 confirmed hvKp cases, there were 50 (80.6%) monomicrobial and 12 (19.4%) polymicrobial infections, whereas among the 88 patients with cKp, there were 43 (46.2%) monomicrobial infections. This difference was significant with P < 0.001.
Antibiotic susceptibility profile of cKp and hvKp
A significant difference in antimicrobial susceptibility patterns was observed between cKp and hvKp. Notably, hvKp exhibited higher susceptibility rates compared to cKp. cKp showed a predominant resistance pattern to fluoroquinolones (levofloxacin and ciprofloxacin), and third-generation cephalosporin (ceftriaxone and ceftazidime), followed by piperacillin-tazobactam, meropenem, and cefoperazonesulbactam. The least resistance percentage was observed for amikacin. Among the hvKp pathotypes, three isolates displayed resistance to all the antibiotics tested. Moreover, MDR was more predominant in cKp (45.5%) compared to hvKp (4.8%), showing a significant difference with P < 0.001. The comparative antibiogram of the isolates is presented in Table 3.
| Antimicrobial agents | cKp (n=88) No. susceptible (%) |
hvKp (n=62) No. susceptible (%) |
P-value |
|---|---|---|---|
| Amikacin (30 µg) | 72 (81.8) | 59 (95.1) | 0.01 |
| Meropenem (10 µg) | 67 (76.1) | 59 (95.1) | 0.01 |
| Piperacillin-tazobactam (100/10 µg) | 65 (73.9) | 59 (95.1) | <0.001 |
| Cefoperazone-sulbactam (75/10 µg) | 67 (76.1) | 59 (95.1) | <0.001 |
| Ciprofloxacin (5 µg) | 26 (30) | 25 (40) | <0.001 |
| Ceftazidime (30 µg) | 56 (63.6) | 59 (95.1) | <0.001 |
| Ceftriaxone (30 µg) | 55 (62.5) | 59 (95.1) | <0.001 |
| Levofloxacin (5 µg) | 25 (28.4) | 34 (54.8) | 0.004 |
| MDR | 23 (45.5) | 3 (4.8) | <0.001 |
P-values were calculated using the Chi-square test or Fisher’s exact test, as appropriate. P-value of ≤ 0.05 is considered as statistically significant and is highlighted in bold. hvKp: Hypervirulent Klebsiella pneumoniae, cKp: Classical K. pneumoniae, MDR: Multidrug resistance.
DISCUSSION
One of the most important traits of K. pneumoniae is its ability to readily acquire foreign genetic material through horizontal gene transfer, enabling it to evolve into a formidable pathogen.[1] At present, two pathotypes are recognized with distinct clinical and epidemiological features. The first pathotype is known as cKp, and the second one is the hvKp pathotype. [7] Different methods are available for the detection of hvKp. Colony morphology and the string test have been considered as screening methods. Other methods, such as the Galleria mellonella infection model, Mouse lethality assay, and Serum killing assay, have been also used; however, these are deemed time-consuming. Recently, several genetic markers emerged as the gold standard for detecting hvKp. Liao et al. stated that the accurate differentiation of hvKp from cKp isolates depended on the presence of peg344 , rmpA, rmpA2, iucA, iron-regulated operon gene B (iroB), and siderophore production.[11] Another study by Russo et al. evaluated the diagnostic accuracy of genes such as rmpA, rmpA1, iucA, iron uptake transporter A (iutA), iroB, peg344, peg589, and peg1631 and assessed their validity. Among all the tested genes, rmpA, iucA, and peg344 exhibited diagnostic accuracies ranging from 96% to 98%.[5] Therefore, these three genes (peg344 , iucA, and rmpA) were selected as the genetic biomarker for hvKp in this study.
In the present study, among the 150 K. pneumoniae isolates tested, 41.3% (62) harbored one or more hypervirulence genes. An isolate carrying at least one of these genes was classified as hvKp. The overall prevalence of hvKp was 41.3%. In an Indian study by Raj et al., it was found that only 11.6% (14/120) of K. pneumoniae isolates were identified as hvKp using the PCR method targeting the iucA gene.[2] Another Indian study by Vandhana et al. reported that 14% (18/129) of K. pneumoniae isolates were classified as hvKp using PCR for the aerobactin gene (iucA).[12] Moreover, both of the Indian studies focused on a single gene for hvKp confirmation. However, in this study, PCR was targeted for three different virulence genes to confirm hvKp. In contrast, Bhardwaj et al. found that only 5.5% (165/3115) had K. pneumoniae infection, of which 19.4% (32/165) of isolates were recognized as hvKp using the phenotypic string test method. The low prevalence could be reliance on the string test alone.[13] The higher prevalence of 41.3% observed in this study is likely due to the targeted inclusion criteria, which focused on infections highly suggestive of the hypervirulent pathotype. The lower prevalence rates reported in many studies were probably due to the absence of such inclusion criteria, with a general pool of K. pneumoniae serving as the denominator.
In the present study, the majority of hypervirulent pathotypes were isolated from males (74.2%), with a mean age of 48 ± 2 years, with a male: female ratio of 2.8:1. Similar to our study, Bhardwaj et al. reported hvKp infection presented in those with a mean age of 53 ± 16.8 years with a male predominance of 73%.[13] A Korean study by Hwang et al. also reported that this hypervirulent pathotype was most commonly encountered in males (69.2%), with a mean age of 68 ± 14 years.[14] The overall majority of the hvKp infection was encountered in males, with a mean age above 50 years. This is in contrast to the earliest studies on hvKp, where a majority was encountered in a younger age group.[15] The reason for the gradual shift in the affected age group is not very clear. There has been an increase in the number of immunocompromised patients in the community setting due to diabetes and chronic kidney disease. This is especially seen in older individuals, whether this has led to the older age group being more affected remains to be investigated. Unfortunately, the comorbidity status of the patients in this study was not documented.
In the present study, the most common clinical presentation of the hypervirulent pathotype was deep-seated abscesses, community-acquired pneumonia, perforation peritonitis, and necrotizing fasciitis. The first hypervirulent pathotype was reported in Taiwan in 1986, and it was associated with PLA.[4] However, they have now become common in invasive infections like bacteremia.[16] hvKp strains have been reported in Asian countries such as India, PR China, South Korea, and Japan, with the initial case documented in Taiwan.[17] This hypervirulent pathotype possesses the unique property of metastatic spread to various sites, and increasing cases of bacteremia due to hvKp infections have been stated in the Russo and Marr study.[1] In this study, abscesses (51.7%) were the most common clinical presentation of hvKp, followed by pneumonia (36.1%) and perforation peritonitis (41.2%). These findings correlate with other published reports. A retrospective study from China reported a maximum number of hvKp from pneumonia (72.3%) followed by invasive infections (18.3%) which include bacteremia and visceral abscesses.[18]
In the present study, among the 56 cases of abscesses, hvKp was isolated from 29 cases. Among them, neck abscesses (6; 10.7%) were the most common clinical presentation of hvKp, followed by splenic abscesses (3; 5.4%) and perinephric abscesses (3; 5.4%). In addition, out of the four isolates tested from liver abscesses, only 3.6% (2/56) were found to carry the virulence genes. In comparison to a study from PR China, where they reported that 10.4% of hvKp infections were associated with liver abscesses and other site abscesses accounted for 26% of cases.[18] Another study from PR China also reported that 16% of hvKp infections were from liver abscesses.[19] In the present study, the majority of samples from neck abscesses were intraoperatively collected pus samples, while a few were FNAC aspirate. However, the possible route of infection spread could not be ascertained.
Conventionally, the string test is the most common rapid phenotypic method used for hvKp identification. However, a recent study by Shankar et al. stated that the string test demonstrated poor sensitivity and specificity and should be no longer considered a valuable method for identifying hvKp.[20] In the present study, the sensitivity and specificity of the string test were found to be 66.7% and 78.5%, respectively, when considering the determination of genetic markers in the plasmid as the gold standard. Among the 150 isolates tested, 62 (41.3%) were positive for virulence genes, and of those, 44 (72%) isolates were string test positive. By comparing it with a study by Sanikhani et al. in Iran, where 477 K. pneumoniae isolates were tested, it was found that 62 isolates (13%) were string test positive, whereas 102 strains (21.4%) harbored virulence genes.[21] Another study by Li et al. reported that out of 495 K. pneumoniae isolates tested, 81 strains (16.4%) had a positive string test, while 71 out of 81 (87.7%) isolates carried virulence genes.[19] The possible reason for the discrepancy between the string test and hypervirulence genes is that the two characteristics, which were earlier considered synonymous, are actually distinct properties, each coded by a different set of genes. An isolate may possess either one alone or both together. Therefore, the current thinking is that the terms “hypermucoviscous” and “hypervirulent” should not be used synonymously.[22,23]
Several virulence genes contribute to the hypervirulent pathotype. This pathotype exhibits a hypermucoviscous phenotype, which is encoded by genes namely, rmpA, rmpA1, iucA, iutA, iroB, peg344, peg589, and peg1631, respectively.[5,15-17] Aerobactin (iucA), a critical siderophore, is produced by more than 90% of hvKp strains.[7] In addition, a divalent metabolic transporter, namely, peg344 enhances the virulence property and its metastatic spread, which is also unique to hvKp.[21] All string test-positive isolates are expected to harbor these virulence genes.
In this study, PCR was carried out for the detection of three virulent genes, including peg344, rmpA, and iucA. Out of 62 hvKp isolates, 55 (88.7%) harbored all three genes, whereas four isolates (6.5%) had iucA alone, two isolates (3.2%) were positive for rmpA alone and only one isolate (1.6%) carried peg344, respectively. The prevalence of iucA gene was higher compared to other genes tested. A study by Du et al. reported that 23.6% (83/352) of K. pneumoniae isolates carried hypervirulence genes. Among the 83 isolates, 42.2% (35/83) had ≥3 genes (iucA, iroB, peg344, rmpA, and rmpA2). The iucA gene was more commonly encountered, as it was detected in 67 isolates (80.7%), whereas rmpA2 was identified in 55 isolates (66.3%). However, only 41% (34/83) of isolates exhibited hypermucoviscosity.[24] In contrast, a study by Sanikhani et al. reported that out of the confirmed 62 hvKp isolates, 45 had iucA alone, six carried iutA alone, while 48 were positive for both iucA and iutA genes and only three isolates exhibited all three genes (iucA, iutA, and peg344).[21] The overall prevalence of iucA, rmpA, and peg344 genes in our study was 92.2%, 92%, and 88.7%, respectively.
In the early stages, the majority of hvKp strains were susceptible to all first- and second-line antimicrobials. However, two mechanisms have been proposed for the establishment of MDR-hvKp strains, one involves the horizontal acquisition of resistance genes through plasmids and mobile genetic elements (MGEs) by hvKp isolates, and the other one involves the acquisition of virulence-associated plasmid (e.g., pLVPK) by MDRcKp isolates. Both pathways eventually result in the establishment of MDR-hvKp strains that are resistant to antibiotics and have high pathogenicity, posing a severe threat to public health. There are a few reports from India quoting the presence of extended-spectrum beta-lactamase (ESBL) and carbapenemase encoding genes in hvKp isolates.[6,12,20,25] In the present study, hypervirulent isolates were found to be significantly more susceptible to antimicrobials compared to classical strains. The highest susceptibility was observed with aminoglycosides, beta-lactam-beta-lactamase inhibitor (BL-BLI) combinations, and carbapenems. Among the cKp tested, the highest susceptibility was seen to aminoglycosides (82%), followed by carbapenems (76%) and BL-BLI combinations (74%). Bhardwaj et al. reported that among 32 confirmed hvKp isolates, 24 (75%) were MDR, while 8 (25%) were sensitive to all the antimicrobials.[13] Another study by Remya et al. revealed that among nine hvKp isolates, seven harbored genes encoding ESBLs such as temoniera (TEM), sulfhydryl variable (SHV), and cefotaximase-munich (CTX-M), whereas, two isolates carried carbapenamse genes, specifically oxacillinase-48 (OXA-48) and new delhi metallo-β-lactamase (NDM).[25] A worrisome feature encountered in our study was the finding of three hvKp isolates that showed resistance to all tested antibiotics. The presentation of this MDR hvKp was infected diabetic foot, multiple oral cavity abscesses, and necrotizing pneumonia. One limitation of this study is that ESBL and carbapenemase-encoding genes were not specifically tested for the three MDR-hvKp isolates. While phenotypic resistance to multiple antimicrobials was observed, the absence of molecular characterization limits the ability to confirm the specific resistance mechanisms involved. Further genetic analysis is needed to better understand the resistance determinants contributing to MDR-hvKp infections.
CONCLUSIONS
The emergence of the hvKp and its subsequent spread across communities in different countries highlights the ease with which this microorganism is able to gain plasmids offering it a survival advantage under a variety of conditions. Further complicating, this scenario is the appearance of resistance plasmids in these isolates converting an already virulent organism into a virtually untreatable one. There is very little data on the true prevalence of this pathotype from India as the simple string test has shown to be unreliable and the ability of most laboratories to perform molecular tests for identification is limited. As this pathotype makes its way into hospitals, it becomes imperative to step up infection control strategies so as to curtail its spread. These include strict implementation of contact precautions and, if feasible, screening of contacts and active surveillance for MDR hvKp, particularly in an outbreak setting.
Author’s contributions
SR: Definition of intellectual content, literature search, clinical studies, experimental studies, data acquisition, data analysis, statistical analysis, manuscript preparation; MM: Definition of intellectual content, literature search, experimental studies, data analysis, manuscript editing and review; MB: Definition of intellectual content, literature search, clinical studies, data analysis, manuscript editing and review; SS: Concepts, design, definition of intellectual content, literature search, clinical studies, data acquisition, data analysis, statistical analysis, manuscript editing and review.
Ethical approval
The research/study was approved by the Institutional Review Board at Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), number JIP/ IEC/2019/332, dated 17th July, 2019.
Declaration of patient consent
Patient consent was not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship: Nil.
References
- Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32:e00001-19.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative analysis of clinical and genomic characteristics of Hypervirulent Klebsiella pneumoniae from hospital and community settings: Experience from a tertiary healthcare center in India. Microbiol Spectr. 2022;10:e0037622.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic surveillance for multidrug-resistant or hypervirulent Klebsiella pneumoniae among United States bloodstream isolates. BMC Infect Dis. 2022;22:603.
- [CrossRef] [PubMed] [Google Scholar]
- Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146:1913-6.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56:e00776-18.
- [CrossRef] [PubMed] [Google Scholar]
- Occurrence and genomic characteristics of hypervirulent Klebsiella pneumoniae in a tertiary care hospital, Eastern India. Infect Drug Resist. 2023;16:2191-201.
- [CrossRef] [PubMed] [Google Scholar]
- Virulence factors in hypervirulent Klebsiella pneumoniae. Front Microbiol. 2021;12:642484.
- [CrossRef] [PubMed] [Google Scholar]
- Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18:94.
- [CrossRef] [PubMed] [Google Scholar]
- Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: Case report and review of molecular mechanisms of pathogenesis. Open Microbiol J. 2011;5:107-13.
- [CrossRef] [PubMed] [Google Scholar]
- Performance standards for antimicrobial susceptibility testing In: CLSI supplement M100 (30th ed). Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- [Google Scholar]
- Rapid detection to differentiate hypervirulent Klebsiella pneumoniae (hvKp) from classical K. pneumoniae by Identifying peg-344 with loop-mediated isothermal amplication (LAMP) Front Microbiol. 2020;11:1189.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of Hypervirulent Klebsiella pneumoniae (HvKp): Correlation of virulence with antimicrobial susceptibility. Int J Microbiol. 2022;2022:4532707.
- [CrossRef] [PubMed] [Google Scholar]
- Incidence, risk factors and clinical outcomes of patients with hypermucoviscoid Klebsiella in a tertiary intensive care unit. J Glob Infect Dis. 2020;12:202-7.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical features and risk factors associated with 30-day mortality in patients with pneumonia caused by hypervirulent Klebsiella pneumoniae (hvKP) Ann Lab Med. 2020;40:481-7.
- [CrossRef] [PubMed] [Google Scholar]
- Hypervirulent (hypermucoviscous) Klebsiella pneumoniae A new and dangerous breed. Virulence. 2013;4:107-18.
- [CrossRef] [PubMed] [Google Scholar]
- Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18:6.
- [CrossRef] [PubMed] [Google Scholar]
- Extensively drug-resistant hypervirulent Klebsiella pneumoniae from a series of neonatal sepsis in a tertiary care hospital, India. Front Med (Lausanne). 2021;8:645955.
- [CrossRef] [PubMed] [Google Scholar]
- High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031-41.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical and microbiological characteristics of invasive and hypervirulent Klebsiella pneumoniae infections in a teaching hospital in China. Infect Drug Resist. 2020;13:4395-403.
- [CrossRef] [PubMed] [Google Scholar]
- Aerobactin seems to be a promising marker compared with unstable rmpA2 for the identification of hypervirulent carbapenem-resistant Klebsiella pneumoniae: In silico and in vitro evidence. Front Cell Infect Microbiol. 2021;11:709681.
- [CrossRef] [PubMed] [Google Scholar]
- The face of hypervirulent Klebsiella pneumoniae isolated from clinical samples of two Iranian teaching hospitals. Ann Clin Microbiol Antimicrob. 2021;20:58.
- [CrossRef] [PubMed] [Google Scholar]
- Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8:1111-23.
- [CrossRef] [PubMed] [Google Scholar]
- Virulence factors and clinical patterns of multiple-clone hypermucoviscous KPC-2 producing K. pneumoniae. Heliyon. 2019;5:e01829.
- [CrossRef] [PubMed] [Google Scholar]
- Nosocomial dissemination of hypervirulent Klebsiella pneumoniae with high-risk clones among children in Shanghai. Front Cell Infect Microbiol. 2022;12:984180.
- [CrossRef] [PubMed] [Google Scholar]
- Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniae. J Lab Physicians. 2018;10:283-8.
- [CrossRef] [PubMed] [Google Scholar]






