Translate this page into:
Evolution of diarrheagenic Escherichia coli pathotypes in India
Address for correspondence: Dr. Shashank Purwar, Department of Medical Microbiology, All India Institute of Medical Sciences, Bhopal - 462 020, Madhya Pradesh, India. E-mail: shashank.microbiology@aiimsbhopal.edu.in
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
CONTEXT:
Diarrheagenic Escherichia coli (DEC) is the leading cause of infectious diarrhea in developing countries. On the basis of virulence and phenotypic characteristics, the DEC is categorized into multiple pathotypes. Each pathotype has different pathogenesis and geographical distribution. Thus, the proper management of disease relies on rapid and accurate identification of DEC pathotypes.
AIMS:
The aim of the study was to determine the prevalence of DEC pathotypes in India.
MATERIALS AND METHODS:
A cross-sectional study was carried out between January 2008 and December 2012 at Jawaharlal Nehru Medical College and KLES Dr. Prabhakar Kore Hospital and Medical Research Center, Belgaum (Karnataka), India. A total of 300 stool samples were collected from diarrhea patients with age >3 months. The DEC was identified by both conventional and molecular methods.
RESULTS:
Of 300 samples, E. coli were detected in 198 (66%) and 170 (56.6%) samples by culture and polymerase chain reaction, respectively. Among DEC (n = 198) isolates, eae gene (59.5%) was the most prevalent followed by stx (27.7%), east (27.2%), elt (12.6%), est (10.6%), ipaH (5.5%), and eagg (1.5%) genes. On the basis of virulence genes, enteropathogenic E. coli (33.8%) was the most common pathotype followed by Shiga toxin-producing E. coli (STEC, 23.2%), enterotoxigenic E. coli (ETEC, 13.6%), enteroinvasive E. coli (5.5%), enteroaggregative heat-stable enterotoxin 1-harboring E. coli (EAST1EC, 4.5%), STEC/ETEC (3.5%), STEC/enteroaggregative E. coli (STEC/EAEC, 1.0%), and EAEC (0.05%).
CONCLUSIONS:
The hybrid DEC is potentially more virulent than basic pathotypes. The pathotyping should be included in clinical settings for the proper management of DEC-associated diarrhea.
Keywords
Enteroaggregative Escherichia coli
enteroaggregative heat-stable enterotoxin 1-harboring Escherichia coli
enteroinvasive Escherichia coli
enteropathogenic Escherichia coli
enterotoxigenic Escherichia coli
Shiga toxin-producing Escherichia coli
Introduction
Infectious diarrhea caused by bacteria, parasites, viruses, and fungi is the leading cause of morbidity and mortality worldwide.[123] Among these pathogens, Escherichia coli (E. coli) is one of the most common causes of bacterial diarrhea in developing countries.[34] On the basis of virulence and phenotypic characteristics, the diarrheagenic E. coli (DEC) are categorized into six main pathotypes including enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), Shiga toxin-producing E. coli (STEC), and diffusely adherent E. coli (DAEC).[5] Each pathotype has a distinct geographical distribution and pathogenesis.[56] Thus, the proper management of the disease depends on the rapid and reliable identification of DEC pathotypes.
Identification and typing both are equally important for the proper management of DEC-associated diarrhea. Today, numerous methods have been developed for the typing/identification of DEC such as serotyping, pulsed-field gel electrophoresis, multilocus satellite typing, multilocus variable-number tandem-repeat analysis, ribotyping, and multiplex polymerase chain reaction (mPCR).[7891011] However, none of these methods can be used for rapid and reliable typing/identification of DEC except mPCR. Moreover, only PCR can differentiate between pathogenic and commensal E. coli.
Several PCR-based studies have been published for the identification of DEC pathotypes.[8121314] However, we cannot neglect the evolution of DEC pathotypes with time because of genomic flexibility of E. coli. In the past few decades, multiple new DEC pathotypes have been reported worldwide.[1516] For instance, the hybrid strains such as STEC/ETEC and STEC/EAEC (O104:H4), these strains cannot be fit into the aforementioned six major pathotypes.[1516] Moreover, the controversy exists regarding the role of EAEC heat-stable enterotoxin 1-harboring E. coli (EAST1EC) in diarrhea.[1718192021] In this study, we used monoplex and multiplex PCRs with minimum set of virulence genes to determine the prevalence of existing and evolving DEC pathotypes.
Materials and Methods
Study population and clinical samples
A cross-sectional study was carried out between January 2008 and December 2012 at Jawaharlal Nehru Medical College and KLES Dr. Prabhakar Kore Hospital and Medical Research Center, Belgaum (Karnataka), India. A total of 300 stool samples were collected from diarrhea patients with age >3 months. Inform consent was obtained from each patient. The study was approved by the Institutional Ethics Committee.
Selection of cases
-
Inclusion criteria – Patients of >3 months of age, suffering from diarrhea
-
Exclusion criteria – Patients of <3 months of age, patients having HIV-associated diarrhea, or patients having diarrhea due administration of antibiotics and cases with already known enteropathogens were excluded from the study.
Isolation and identification of Escherichia coli
Stool samples were cultured on MacConkey agar and incubated at 37°C for 18-24 hours. The lactose-fermenting colonies were subjected to conventional and molecular identification. The samples positive for one of the enteric pathogens, namely Salmonella spp., Shigella spp., Vibrio cholerae, Candida, and trophozoites/cyst/ova of the parasites were excluded from the study.
Bacterial strains
The control strains of EPEC, ETEC, EIEC, enterohemorrhagic E. coli (EHEC), and EAEC were provided by Dr. Shanta Dutta, Scientist “F,” of the National Institute of Cholera and Enteric Diseases, Kolkata. The reference strains were positive for virulence genes included in PCR, namely EPEC: eae gene, ETEC: elt and est genes, STEC: stx gene, EIEC: ipaH gene, and EAEC: eagg gene.
DNA extraction
DNA extraction was carried out using the cetyltrimethylammonium bromide (CTAB) method.[22] In brief, the culture was centrifuged at 8000 revolution per minute (rpm) for 5 minutes (min), and the supernatant was discarded and transferred the pellet to a vial containing 10 mg/mL lysozyme (Fermentas Inc., Maryland, USA). Following lysis of bacterial cells, the lysate treated with 10% sodium dodecyl sulfate (56 μL) and proteinase K (5 μL) (Fermentas Inc., Maryland, USA). For polysaccharide precipitation, 5 M sodium chloride (NaCl) and CTAB/NaCl (64 μL) (Fermentas Inc., Maryland, USA) were added and incubated at 65°C for 30 min. After this, an equal amount of chloroform: isoamyl alcohol solution (Fermentas Inc., Maryland, USA) was added to the mix and centrifuged at 10,000 rpm for 15 min followed by the precipitation of DNA with ethanol. Finally, the DNA precipitate was resuspended in 40 μL Tris-EDTA (TE) buffer.
Polymerase chain reaction
The primer sequences used for DEC pathotype identification were retrieved from the previous publication.[2324252627] The primers were synthesized by Sigma (Sigma-Aldrich Co., St. Louis, MO, USA) and specificity for targeted genes (eae, astA, eagg, stx, ipaH, elt, and est genes) confirmed by monoplex PCR [Supplementary Table 1]. The multiplex PCR assays were standardized by testing different PCR cycling conditions. The criteria used for the identification of DEC pathotypes are enlisted in Supplementary Table 2. The two pairs of primers were used in each mPCR because of non-specific amplification and primer-dimer formations; therefore, five multiplex PCRs were standardized for the identification of DEC.
| Gene | Primer sequence (5`-3`) | Amplicon size (bp) | Annealing temperature (°C) | Reference number |
|---|---|---|---|---|
| eae | AAACAGGTGAAACTGTTGCC CTCTGCAGATTAACCCTCTGC |
454 | 54 | 24 |
| elt | GCGACAAATTATACCGTGCT CCGAATTCTGTTATATATATGT |
708 | 55 | 24 |
| est | ATTTTTA/CTTTCTGTATTA/GTCTT CACCCGGTACAA/GGCAGGATT |
190 | 55 | 24 |
| stx | CAG TTA ATG TGG TTG CGA AG CTG CTA ATA GTT CTG CGC ATC |
895 | 55 | 25 |
| ipaH | TGG AAA AAC TCA GTG CCT CT CCA GTC CGT AAA TTC ATT CT |
423 | 60 | 27 |
| eagg | CTGGCGAAAGACTGTATCAT CAATGTATAGAAATCCGCTGTT |
630 | 54 | 23 |
| astA | CACAGTATATCCGAAGGC CGAGTGACGGCTTTGTAG |
94 | 55 | 24 |
| aggR | CTAATTGTACAATCGATGTA ATGAAGTAATTCTTGAAT |
308 | 55 | 26 |
| aap | CTTTTCTGGCATCTTGGGT GTAACAACCCCTTTGGAAGT |
232 | 54 | 26 |
| shf | ACTTTCTCCCGAGACATTC CTTTAGCGGGAGCATTCAT |
613 | 53 | 26 |
| Criteria (gene detected) | Interpretation |
|---|---|
| eae and/or astA but no stx/est/elt/ipaH/eagg | EPEC |
| eae and/or stx and/or astA | STEC |
| est and/or elt and/astA | ETEC |
| ipaH and/or astA | EIEC |
| eagg and/or astA | EAEC |
| stx and elt/est and/or astA | STEC/ETEC |
| stx and eagg and/or astA | STEC/EAEC |
| None of the above genes detected but only astA | EAST1EC |
| None of the above genes detected | DECNVG |
E. coli=Escherichia coli, EPEC=Enteropathogenic E. coli, ETEC=Enterotoxigenic E. coli, EIEC=Enteroinvasive E. coli, EAEC=Enteroaggregative E. coli, STEC=Shiga toxin-producing E. coli, STEC/EAEC=E. coli with stx and eagg genes, STEC/ETEC=E. coli with stx and elt/est or both genes, EAST1EC=EAEC heat-stable enterotoxin 1-harboring E. coli (possess no other diarrheagenic characteristics), DECNVG=Diarrheagenic Escherichia coli with no virulence genes
-
Multiplex 1: Identification EPEC (eae and astA genes)
-
Multiplex 2: Identification ETEC (elt and astA genes)
-
Multiplex 3: Identification ETEC (est and astA genes)
-
Multiplex 4: Identification STEC (stx and astA genes)
-
Multiplex 5: Identification EAEC (eagg and astA genes).
Each monoplex and multiplex PCRs were performed using 25 μL reaction mixtures. The master mix included 10X PCR buffer with MgCl2: 2.5, dNTP mix: 2.0 μL (200 μM each), forward: 1.0 μL (400 nM), reverse primer: 1.0 μL (400 nM), template DNA: 2.5 μL (100 ng), sterile water: 15.8 μL, and Taq polymerase: 0.2 μL (0.4 U) (Fermentas Inc., Maryland, USA). For multiplex PCRs, the volume of water was adjusted, according to the number of primers. The PCR thermocycling conditions were as follows: initial denaturation 94°C for 5 min followed by 35 cycles of denaturation (94°C for 30 seconds), annealing (primer dependent), extension (72°C for 30 seconds), and final extension (72°C for 10 min).[9]
Statistical analysis
All statistical analysis was done using Statistical Package for the Social Sciences (SPSS) for Windows version 22.0 (IBM Corp., Armonk, New York, USA). The patients were classified into multiple groups according to their age.
Results
Out of 300, 102 (34%) samples were positive for one of the enteric pathogens and the remaining 198 (66%) samples yielded growth of E. coli [Table 1]. Among E. coli (n = 198) isolates, PCR detected virulence genes in 170 (85.8%) isolates only. In DEC pathotypes, EPEC (33.8%) was the predominant pathotypes followed by STEC (23.2%), ETEC (13.6%), EIEC (5.5%), EAST1EC (4.5%), EIEC with east (1.7%), STEC/ETEC (3.5%), and STEC/EAEC (1.0%). In about 14.1% DEC, no virulence genes were detected [Figure 1].
| Serial number | Pathogens isolated/observed | Number of cases (%) | Remarks (%) |
|---|---|---|---|
| 1 | E. coli | 198 (66) | |
| 2 | Other pathogens | ||
| A | Other bacteria (Vibrio cholerae, Salmonella spp., and Shigella spp.) | 36 (12) | 102 (34) cases was not associated with DEC |
| B | Parasites (protozoan parasite and helminths) | 49 (16.3) | |
| C | Candida spp. (Candida was presumed as causative pathogen on the basis of the presence of large number of budding yeasts resembling Candida in morphology in the stool samples and the absence of any other known diarrheagenic pathogen. The Candida isolates were differentiated by germ tube test into albicans and non-albicans spp. All isolates were C. albicans) | 17 (5.6) | |
| Total | 300 |
E. coli=Escherichia coli, C. albicans=Candida albicans
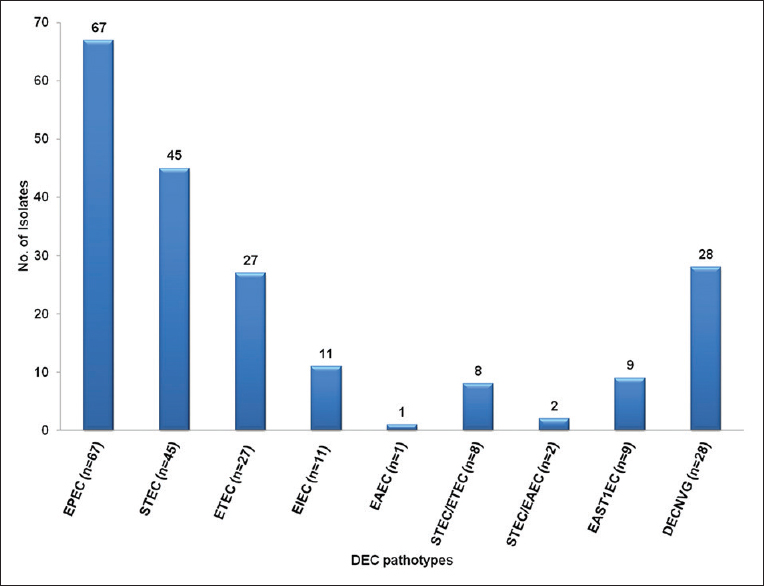
- Prevalence of DEC pathotypes (n = 198). E. coli = Escherichia coli, DEC = Diarrheagenic Escherichia coli, EPEC = Enteropathogenic Escherichia coli, ETEC = Enterotoxigenic Escherichia coli, EIEC = Enteroinvasive Escherichia coli, EAEC = Enteroaggregative Escherichia coli, STEC = Shiga toxin-producing Escherichia coli, STEC/EAEC = Escherichia coli with stx and eagg genes, STEC/ETEC = Escherichia coli with stx and elt/est or both genes, EAST1EC = EAEC heat-stable enterotoxin 1-harboring Escherichia coli (possess no other diarrheagenic characteristics), DECNVG = Diarrheagenic Escherichia coli with no virulence genes
In E. coli (n = 198), the eae gene (59.5%) was the most prevalent followed by stx (27.7%), east (27.2%), elt (12.6%), est (10.6%), ipaH (5.5%), and eagg (1.5%) [Figure 2]. The overlapping of virulence genes was observed among DEC pathotypes. The east gene was detected in all DEC pathotypes [Figure 2].

- Distribution of virulence genes among DEC (n = 198), E. coli = Escherichia coli, DEC = Diarrheagenic Escherichia coli, EPEC = Enteropathogenic Escherichia coli, ETEC = Enterotoxigenic Escherichia coli, EIEC = Enteroinvasive Escherichia coli, EAEC = Enteroaggregative Escherichia coli, STEC = Shiga toxin-producing Escherichia coli, STEC/EAEC = Escherichia coli with stx and eagg genes, STEC/ETEC = Escherichia coli with stx and elt/est or both genes, EAST1EC = EAEC heat-stable enterotoxin 1-harboring Escherichia coli (possess no other diarrheagenic characteristics), DECNVG = Diarrheagenic Escherichia coli with no virulence genes
The age of patients ranges from 4 months to 40 years. Eighty-three (n = 83, 41.9%) patients were <5 years of age and 115 patients (n = 115, 58%) were >5 years of age. Among different age groups, the maximum number of diarrhea cases was seen in children [Figure 3].

- Distribution of DEC pathotypes among different age groups (n = 170), E. coli = Escherichia coli, DEC = Diarrheagenic Escherichia coli, EPEC = Enteropathogenic Escherichia coli, ETEC = Enterotoxigenic Escherichia coli, EIEC = Enteroinvasive Escherichia coli, EAEC = Enteroaggregative Escherichia coli, STEC = Shiga toxin-producing Escherichia coli, STEC/EAEC = Escherichia coli with stx and eagg genes, STEC/ETEC = Escherichia coli with stx and elt/est or both genes, EAST1EC = EAEC heat-stable enterotoxin 1-harboring E. coli (possess no other diarrheagenic characteristics), DECNVG = Diarrheagenic Escherichia coli with no virulence genes
Discussion
DEC-associated outbreaks have risen in advanced and developing nations in recent decades, recognizing the significance of DEC globally.[56] In India, multiple studies have been published on the pediatric population but very few targeted adults.[123] Thus, we included both children and adults in this study. The prevalence of DEC in our study was much higher (66%) than previous studies.[23] The inclusion of the adult age group may be the reason of higher prevalence. On comparing the children and adults, the low prevalence of DEC was seen in adult. The low prevalence of DEC in adults may also be due to mucosal immunity, loss of receptors, and acquired immunity.[5]
Among DEC pathotypes, EPEC (33.8%, 67/198) was the most common pathotype, corresponding to the studies from other developing countries. Although EPEC frequently isolated from persistent infection in children, we detected the cases in both children and adults, suggesting that EPEC is not limited to children only. On the basis of virulence genes (eae and bfp), EPEC is classified into typical (eae) and atypical EPEC (bfp).[6] In atypical EPEC, multiple alleles of bfp gene exist, which cannot be targeted by single PCR.[28] Thus, we targeted only typical EPEC in this study.
Verocytotoxigenic E. coli/STEC/Shiga-like toxin-producing E. coli/EHEC represents one group of DEC.[5] The STEC terminology was used in this study. Most of the studies reported the low prevalence of STEC in India, whereas we observed the high prevalence of STEC (46, 23.2%), suggesting poor hygiene and sanitation in this region.[6] ETEC is characterized by the expression of elt or est gene only or both.[56] The isolates which possessed either of elt/est or both the genes were identified as ETEC pathotypes in this study. The prevalence of ETEC varies from 10% to 30%.[2930] Similarly, we observed a 13% prevalence in this study.
EIEC is characterized by ipaH gene expression.[531] However, ipaH gene sequence cannot differentiate between Shigella and EIEC.[5] The EIEC isolates may also be missed or misidentified as Shigella species by this PCR. To exclude the Shigella, only lactose-fermenting, indole-producing E. coli colonies were subjected to PCR in this study. Despite this, the possibility of missing some lactose negative, nonmotile, and lysine decarboxylase-negative EIEC strains cannot be rule out. In addition, EIEC are missed in clinical laboratory because of suspecting them only in stool sample with mucus and blood, suggesting that the actual incidence of EIEC may also be slightly higher than the result derived in our study.[5]
EAEC is characterized by the absence of heat-labile/heat-stable enterotoxin and the presence of aggregative adherence.[1732] The eagg, aap, aggR, and shf genes were targeted for identification of EAEC on the basis of previous studies.[2326] EAEC is considered as emerging DEC pathotypes.[2633] However, a very low prevalence of EAEC was found in this study. In addition, stx gene was also detected in two EAEC (n = 3). Hence, we cannot draw any conclusion regarding the pathogenicity of EAEC with this small number of isolates.
Among DEC, the astA gene (EAST1 toxin) has been reported in multiple DEC pathotypes: ETEC, STEC, and EPEC.[1819202134] Similarly, the astA gene expression was detected in EPEC, ETEC, STEC, EIEC, and EAEC in this study. Moreover, we also identified the nine isolates (4.5%) with astA gene only, correlating with the findings of Nishikawa et al. 1999 and 2002.[3536] This suggests that some unrecognized DEC pathotypes exist, which can cause diarrhea even in the absence of typical virulence factors. However, few studies reported no role of astA gene in diarrhea.[1920] Further, the study with large number of isolates may provide insight within the exact role of astA gene in diarrhea.
In the past few decades, multiple hybrid strains of DEC have been reported worldwide.[15163738] Similarly, we also detected STEC/ETEC (7, 2.3%) and STEC/EAEC (2, 0.6%) in this study. The high prevalence of STEC may also be the reason of emergence of hybrid strains. The hybrid strains contain multiple virulence genes, making them more virulent and pathogenic.[3738] The emergence of these hybrid strains may pose a serious threat to public health.
Among DEC, 28 (14.1%) isolates were with no known diarrhea-associated virulence genes, suggesting the existence of some unknown virulence genes and DEC pathotypes, which cannot be detected by targeting common virulence genes such as DAEC and cell-detaching E. coli.[39] Further studies may define the role of these DEC in diarrhea.
Conclusions
In brief, the DEC is evolving in India, indicated by the presence of hybrid strain [Figure 4]. The combination of phenotypic and genotypic methods should be used in clinical setting for the reliable identification of typical and emerging DEC pathotypes. Moreover, the surveillance system should be established to monitor DEC-associated outbreak.

- Model for overlapping virulence genes among DEC pathotypes. E. coli = Escherichia coli, DEC = Diarrheagenic Escherichia coli, EPEC = Enteropathogenic Escherichia coli, ETEC = Enterotoxigenic Escherichia coli, EIEC = Enteroinvasive Escherichia coli, EAEC = Enteroaggregative Escherichia coli, STEC = Shiga toxin-producing Escherichia coli, STEC/EAEC = Escherichia coli with stx and eagg genes, STEC/ETEC = Escherichia coli with stx and elt/est or both genes, EAST1EC = EAEC heat-stable enterotoxin 1-harboring Escherichia coli (possess no other diarrheagenic characteristics), DECNVG = Diarrheagenic Escherichia coli with no virulence genes
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- The global enteric multicenter study (GEMS) of diarrheal disease in infants and young children in developing countries: Epidemiologic and clinical methods of the case/control study. Clin Infect Dis. 2012;55(Suppl 4):S232-45.
- [Google Scholar]
- Aetiology of acute paediatric gastroenteritis in Bulgaria during summer months: Prevalence of viral infections. J Med Microbiol. 2015;64:272-82.
- [Google Scholar]
- Multiple etiologies of infectious diarrhea and concurrent infections in a pediatric outpatient-based screening study in Odisha, India. Gut Pathog. 2017;9:16.
- [Google Scholar]
- Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol. 2015;64:432-7.
- [Google Scholar]
- Characterisation of diarrhoeagenic Escherichia coli clones by ribotyping and ERIC-PCR. J Med Microbiol. 1998;47:227-34.
- [Google Scholar]
- Detection of diarrheagenic Escherichia coli by multiplex PCR. Indian J Med Microbiol. 2012;30:279-84.
- [Google Scholar]
- Genotyping of ESBL producing uropathogenic and diarrheagenic Escherichia coli in southeast of Iran. Infect Disord Drug Targets. 2015;15:118-24.
- [Google Scholar]
- Virulence gene profile and multilocus variable-number tandem-repeat analysis (MLVA) of enteroinvasive Escherichia coli (EIEC) isolates from patients with diarrhea in Kerman, Iran. Jundishapur J Microbiol. 2016;9:e33529.
- [Google Scholar]
- Prevalence and genetic diversity of human diarrheagenic Escherichia coli isolates by multilocus sequence typing. Int J Infect Dis. 2018;67:7-13.
- [Google Scholar]
- Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol. 2003;41:2669-71.
- [Google Scholar]
- Rapid categorization of pathogenic Escherichia coli by multiplex PCR. Microbiol Immunol. 2005;49:485-92.
- [Google Scholar]
- A novel single-step multiplex polymerase chain reaction assay for the detection of diarrheagenic Escherichia coli. J Microbiol Meth. 2013;92:289-92.
- [Google Scholar]
- Characteristics of the shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int Microbiol. 2011;14:121-41.
- [Google Scholar]
- Comparative genomics and characterization of hybrid shigatoxigenic and enterotoxigenic Escherichia coli (STEC/ETEC) strains. PLoS One. 2015;10:e0135936.
- [Google Scholar]
- Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J Clin Invest. 1991;87:1450-55.
- [Google Scholar]
- Comparison of the nucleotide sequence of enteroaggregative Escherichia coli heat-stable enterotoxin 1 genes among diarrhea-associated Escherichia coli. FEMS Microbiol Lett. 1997;147:89-95.
- [Google Scholar]
- Escherichia coli expressing EAST1 toxin did not cause an increase of cAMP or cGMP levels in cells, and no diarrhea in 5-day old gnotobiotic pigs. PloS one. 2012;7:e43203.
- [Google Scholar]
- Detection of virulence factors of Escherichia coli focused on prevalence of EAST1 toxin in stool of diarrheic and non-diarrheic piglets and presence of adhesion involving virulence factors in astA positive strains. Vet Microbiol. 2012;154:369-75.
- [Google Scholar]
- Detection and genetic analysis of the enteroaggregative Escherichia coli heat-stable enterotoxin (EAST1) gene in clinical isolates of enteropathogenic Escherichia coli (EPEC) strains. BMC Microbiol. 2014;14:135.
- [Google Scholar]
- Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett. 2011;325:162-9.
- [Google Scholar]
- Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701-5.
- [Google Scholar]
- Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J Clin Microbiol. 2001;39:3241-6.
- [Google Scholar]
- Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol. 2002;51:548-56.
- [Google Scholar]
- Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2004;42:4111-20.
- [Google Scholar]
- Rapid detection of diarrheagenic E. coli by real-time PCR. J Microbiol Methods. 2005;61:335-41.
- [Google Scholar]
- Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect Immun. 2000;68:7028-38.
- [Google Scholar]
- Molecular characterization of diarrheagenic Escherichia coli pathotypes: Association of virulent genes, serogroups, and antibiotic resistance among moderate-to-severe diarrhea patients. J Clin Lab Anal. 2018;32:e22388.
- [Google Scholar]
- Diarrhoeagenic Escherichia coli as a predominant cause of paediatric nosocomial diarrhoea in India. J Med Microbiol. 2012;61:830-6.
- [Google Scholar]
- Detection of shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J Infect Dis. 1993;167:458-61.
- [Google Scholar]
- Autotransporters and virulence of enteroaggregative E. coli. Gut Microbes. 2011;2:13-24.
- [Google Scholar]
- Prevalence of enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland. J Clin Microbiol. 2003;41:2289-93.
- [Google Scholar]
- Occurrence of virulence genes associated with Diarrheagenic pathotypes in Escherichia coli isolates from surface water. Applied and environmental microbiology. 2013;79:328-35.
- [Google Scholar]
- An outbreak of gastroenteritis in Japan due to Escherichia coli O166. Emerg Infect Dis. 1999;5:300.
- [Google Scholar]
- Diarrheagenic Escherichia coli isolated from stools of sporadic cases of diarrheal illness in Osaka city, Japan between 1997 and 2000: Prevalence of enteroaggregative E. coli heat-stable enterotoxin 1 gene-possessing E. coli. Jpn J Infect Dis. 2002;55:183-90.
- [Google Scholar]
- Heterogenic virulence in a diarrheagenic Escherichia coli: Evidence for an EPEC expressing heat-labile toxin of ETEC. Int J Med Microbiol. 2015;305:47-54.
- [Google Scholar]
- Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709-17.
- [Google Scholar]
- Diffuse and enteroaggregative patterns of adherence of enteric escherichia coli isolated from aboriginal children from the Kimberley region of Western Australia. J Infect Dis. 1993;167:755-8.
- [Google Scholar]





