Translate this page into:
Identification of genus Acinetobacter: Standardization of in-house PCR and its comparison with conventional phenotypic methods
Address for correspondence: Dr. Raghavendra D. Kulkarni, Department of Microbiology, SDM College of Medical Sciences and Hospital, Sattur, Dharwad - 580 009, Karnataka, India. E-mail: atul410@yahoo.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
Acinetobacter is grouped under nonfermenting Gram-negative bacilli. It is increasingly isolated from pathological samples. The ability of this genus to acquire drug resistance and spread in the hospital settings is posing a grave problem in healthcare. Specific treatment protocols are advocated for Acinetobacter infections. Hence, rapid identification and drug susceptibility profiling are critical in the management of these infections.
AIMS:
To standardize an in-house polymerase chain reaction (PCR) for identification of genus Acinetobacter and to compare PCR with two protocols for its phenotypic identification.
METHODOLOGY:
A total of 96 clinical isolates of Acinetobacter were included in the study. An in-house PCR for genus level identification of Acinetobacter was standardized. All the isolates were phenotypically identified by two protocols. The results of PCR and phenotypic identification protocols were compared.
RESULTS:
The in-house PCR standardized was highly sensitive and specific for the genus Acinetobacter. There was 100% agreement between the phenotypic and molecular identification of the genus. The preliminary identification tests routinely used in clinical laboratories were also in complete agreement with phenotypic and molecular identification.
CONCLUSION:
The in-house PCR for genus level identification is specific and sensitive. However, it may not be essential for routine identification as the preliminary phenotypic identification tests used in the clinical laboratory reliably identify the genus Acinetobacter.
Keywords
Acinetobacter
nonfermenting Gram-negative bacilli
polymerase chain reaction
Introduction
Acinetobacter is a common isolate, especially in the hospital setting. Of late, the isolation of this pathogen has become a grave problem in the Intensive Care Units (ICUs). In fact, it is the second most common nonfermenting Gram-negative bacilli (NFGNB), causing bacterial infections after Pseudomonas. Acinetobacter is grouped under NFGNB. Its ability to acquire drug resistance has become a difficult problem, and recovery of pan-resistant Acinetobacter is not uncommon. Acinetobacter is oxidase negative, nonmotile, nonfermenting, Gram-negative, coccobacillus. Brison and Prevot (1954) proposed the generic designation, Acinetobacter. In 1971, the Subcommittee on Taxonomy of Moraxella and allied bacteria suggested that the genus Acinetobacter shall include only oxidase negative, nonmotile, nonfermenting, Gram-negative coccobacillus.[12]
At present, 33 genomospecies of Acinetobacter have been recognized by DNA–DNA hybridization. Among these species, Acinetobacter calcoaceticus, Acinetobacter baumannii, Acinetobacter genomic species 3, and Acinetobacter genomic species 13TU are very closely related and are difficult to distinguish from each other phenotypically. They are said to be saccharolytic strains. Therefore, they have been grouped as A. calcoaceticus-A. baumannii complex. This group accounts for 80% of the clinical infections caused by Acinetobacter spp. A. baumannii is the most frequently isolated species from human clinical specimens, followed by Acinetobacter spp. 3. Androstachys johnsonii, Acinetobacter lwoffii, and Acinetobacter spp. 12, which are nonsaccharolytic occur as natural inhabitants of human skin.[2]
Molecular methods such as amplified 16S rRNA gene restriction analysis, high-resolution fingerprint analysis by amplified fragment length polymorphism, ribotyping, tRNA spacer fingerprinting, restriction analysis of the 16S–23S rRNA intergenic spacer sequences, and sequence analysis of the 16S–23S rRNA gene spacer region are in use for the identification of Acinetobacter. These methods are used in only a few reference laboratories and are not suitable for use in routine laboratories. Gundi et al. have shown that partial rpo B gene sequence analysis is a simple molecular tool for the identification and speciation of Acinetobacter clinical strains.[23]
We discuss here the development of in-house polymerase chain reaction (PCR) and its sensitivity and specificity for genus level identification of Acinetobacter. We also compared the accuracy of identification of genus Acinetobacter based on preliminary tests with phenotypic and molecular identification.
Methodology
The study was conducted in the Department of Microbiology of a tertiary care center in North Karnataka. A total of 96 Acinetobacter isolates from various clinical samples such as endotracheal (ET) secretions, ET tubes, sputum, pus, blood, and cerebrospinal fluid and environmental isolates from surveillance cultures of the medical ICU were also included in this work. All the samples were processed according to the standard laboratory techniques. Three identification protocols, viz., identification based on preliminary biochemical tests, identification using standard biochemical and physiological tests, and PCR were used for the identification of the isolates.
The preliminary identification tests after the plate reading were Gram's stain, catalase test, oxidase test, and motility. Further identification was continued by standard biochemical and physiological tests.[4567] All the isolates were also subjected to PCR, targeting 350 bp hypervariable region of rpoB gene.
Polymerase chain reaction
We used ATCC A. baumannii 19606 as reference strain for standardization of the in-house PCR. The strain was subcultured on brain–heart infusion (BHI) agar plate to check for purity. It was phenotypically reconfirmed. Two well-characterized colonies from BHI agar were inoculated into 1 ml of LuriaBertani (LB) broth and incubated overnight at 37°C. Extraction of DNA was carried out by phenol–chloroform method. The amplification was carried out using primers (Ac696FAc1093R) procured from Sigma-Aldrich, targeting 350 bp hypervariable zone in the rpo B gene specific to Acinetobacter species.[3] The PCR mixture (50 μl) contained 2 μl DNA, PCR Master–Mix (2X) (Chromous Biotech), and primers at 0.2 μM concentration. The protocol for amplification used was initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s and extension at 72°C for 1 min followed by final elongation at 72°C for 7 min. The PCR product was resolved on 1% agarose gel containing ethidium bromide in Tris-acetate ethylenediaminetetraacetic acid buffer at 100 V for 1 h. The amplicon was examined and documented using gel doc system (Zenith Research, Mumbai).
The sensitivity and specificity of the in-house PCR were determined by harvesting growth from LB broths. LB broths were inoculated and incubated at 37°C for 24 h, and pellet was obtained by centrifugation at 3000 rpm for 5 min. The pellet was washed and resuspended in sterile nuclease free distilled water to obtain doubling dilutions from 1:2 to 1:128. DNA was extracted from these diluted samples by phenol–chloroform extraction method and subjected to PCR. The DNA concentration of the all the dilutions was estimated using Epoch™, Biotek spectrophotometer.
For determining the specificity, we subjected various routine bacterial isolates recovered in our bacteriology laboratory to this PCR. We tested Staphylococcus aureus (n = 4), Enterococcus spp. (n = 4), Streptococcus spp. (n = 4), Moraxella spp. (n = 4), Klebsiella pneumoniae (n = 4), Escherichia coli (n = 4), Pseudomonas aeruginosa (n = 4), Salmonella Enterica Typhi (n = 4), Providencia rettgeri (n = 4) NFGNB including Stenotrophomonas (n = 6). We used ATCC A. baumannii 19606 and well-characterized stock strains of A. lwoffii and A. calcoaceticus from our laboratory as positive controls. The PCR amplified all Acinetobacter whereas it did not amplify any of the non-Acinetobacter isolates.
A total of 96 Acinetobacter clinical isolates were subjected to the standardized PCR. One amplicon from a positive PCR reaction was randomly selected and sent for sequencing (Chromous Biotech, Bangalore) and analyzed by basic local alignment search tool which confirmed it to be belonging to the genus Acinetobacter.
Results
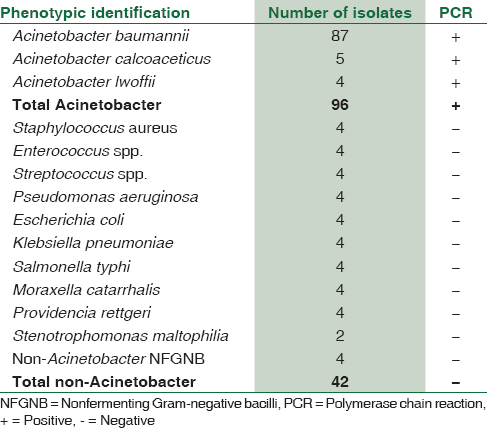
Of the 96 Acinetobacter spp. included in the study, 87 were identified as A. baumannii, 5 as A. calcoaceticus, and 4 as A. lwoffii by standard biochemical and physiological tests. All the isolates were identified as Acinetobacter spp. by the preliminary tests, viz., Gram stain, catalase test, oxidase test, and motility by hanging drop preparation. All the 96 isolates successfully amplified the requisite target gene [Table 1].

A set of 42 non-Acinetobacter isolates [Table 2] recovered from various samples were also subjected to the same PCR assay. All these 42 isolates did not show amplification for the target gene. Thus, there was 100% agreement between the preliminary phenotypic identification and PCR for genus level identification of Acinetobacter [Table 1].
The overnight incubated LB broth of ATCC A. baumannii 19606 was diluted by doubling dilutions in distilled water and was subjected to PCR. The highest dilution that gave a positive amplification was 1:64. The DNA extracted from this dilution on quantitation by Epoch™, Biotek Spectrophotometer showed a concentration of 8.426 ng/μl.
Discussion
Acinetobacter is the second most common NFGNB isolated in the clinical laboratories. Acinetobacter infection has emerged as a serious threat to the health-care system because of its multidrug resistance drifting to pan-resistance. Various mechanisms such as β-lactamase production, alterations in cell wall channels (porins), efflux pumps, and carbapenemase production have been recognized conferring resistance to this pathogen. Strains producing metallo-β-lactamase are frequently encountered. Therapy for carbapenem-resistant Acinetobacter is a serious problem. The current available choices such as polymyxins or tigecycline are also under threat of drug resistance.[89]
Acinetobacter is frequent colonizer of the hospital personnel, patients, and environment. Hands of the health-care workers, respiratory tract and skin of the patients, inanimate objects such as floor, bed, and computer keyboards are often colonized.[8] Accurate identification and speciation of this organism are important for successful treatment as well as for epidemiological studies.
Many clinical laboratories stop at genus level identification of the Acinetobacter and do not go ahead with further biochemical analysis. The identification is mostly based on preliminary tests such as Gram's stain, catalase test, oxidase test, and hanging drop preparation for motility. An important test such as nitrate reduction test is also not used. As per the standard norms, the identification of any pathogen based on preliminary tests is not very reliable. Other NFGNBs such as Burkholderia spp. and Stenotrophomonas spp. are isolated frequently in clinical laboratories and the phenotypic identification of these organisms is uncertain. Identification of NFGNBs, giving the oxidase test negative, is an enigma for clinical laboratories. We, therefore, used an in-house PCR targeting a 350 bp rpo B gene for genus level identification of Acinetobacter. This PCR has been shown to be specific for identification of Acinetobacter.[3] A total of 98 clinical isolates (including stock strains as internal control) and ATCC Acinetobacter 19606 strains of Acinetobacter [Table 2] were subjected to the in-house PCR. The clinical isolates were speciated by phenotypic tests. Of the 96 clinical isolates, 87 were A. baumannii, 5 were A. calcoaceticus, and 4 were A. lwoffii. All the phenotypically identified Acinetobacter gave amplification for the requisite target. Thus, there was 100% agreement between phenotypic identification and molecular identification of Acinetobacter. When 42 non-Acinetobacter bacterial isolates were subjected to this PCR, none of the isolates gave amplification for the target gene. This showed that the in-house PCR was highly specific for the genus Acinetobacter.
We tried to correlate the identification of Acinetobacter by preliminary tests (Gram stain, catalase, oxidase, and motility) and in-house PCR. It was observed that there was complete agreement between the preliminary identification and PCR [Table 1]. This is highly assuring for the clinical laboratories practicing identification of Acinetobacter based on preliminary tests. This shows that the preliminary tests commonly used by clinical laboratories for identification of Acinetobacter are reliable. Considering the importance of Acinetobacter in human infections, rapid identification of this organism is of paramount importance in treatment initiation and further management of the patients. A lot of clinical laboratories including tertiary care hospitals are still not using PCR routinely for logistic and financial constraints. This study indicates that the basic preliminary test protocols when implemented meticulously identify genus Acinetobacter precisely. This PCR assay needs to be further standardized for detection of Acinetobacter with its drug resistance profile directly from clinical specimens, which will be a boon to hasten the patient care. This assay also may also have a prominent role in the infection control practices.
The three most clinically relevant Acinetobacter species are A. baumannii, Acinetobacter nosocomialis (formerly Acinetobacter genomic species 13TU), and Acinetobacter pittii (formerly Acinetobacter genomic species 3).[10] However, these cannot be differentiated by phenotypic tests including the automated identification systems such as API 20NE, Vitek2, Phoenix, and MicroScan WalkAway systems. Molecular methods such as the detection of blaOXA51like, the intrinsic carbapenemase gene in A. baumannii, and sequencing of the rpoB gene have helped to identify and speciate the genus. A. baumannii is the most resistance one of the genomospecies and has substantial clinical relevance. This pathogen is the most frequently isolated species and is typically associated with outbreaks in the hospital setting.[2] It is endowed with resistance to harsh environmental factors, enabling it to establish and spread rapidly in the hospital environment.[111213]
Considering the increasing threat of this highly drug-resistant pathogen as a hospital as well as community pathogen, it is very important to identify it quickly and accurately as Acinetobacter. Rapid identification will be important for therapeutic as well as epidemiological needs to identify and check the spread of this pathogen. Immediate implementation of disinfection methods and alarming the hospital personnel about the presence of this pathogen will help mitigate the menace.
Conclusion
The phenotypic identification based on preliminary tests commonly practiced in clinical laboratories has excellent correlation with the molecular identification. Accurate identification of genus Acinetobacter can be performed using preliminary tests, viz., Gram stain, catalase, oxidase, and motility, which can be easily performed in any clinical laboratory. In-house PCR standardized in our study is specific and sensitive for the identification of Acinetobacter species. There is a need to standardize this PCR assay for direct identification of Acinetobacter from the clinical specimens.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Koneman's Color Atlas and Textbook of Diagnostic Microbiology (6th ed). Philadelphia, USA: Lippincott Williams and Wilkins; 2006. p. :353-5. Ch. 7
- Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538-82.
- [Google Scholar]
- Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009;155:2333-41.
- [Google Scholar]
- Taxonomy of the Genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov. Acinetobacter haemolyticus sp. nov. Acinetobacter johnsonii sp. nov. and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwofii. Int J Syst Bacteriol. 1986;36:228-40.
- [Google Scholar]
- Clinical and antimicrobial profile of Acinetobacter spp.: An emerging nosocomial superbug. Adv Biomed Res. 2014;3:13.
- [Google Scholar]
- Identification and speciation of Acinetobacter and their antimicrobial susceptibility testing. Al Ameen J Med Sci. 2010;3:345-9.
- [Google Scholar]
- Multidrug-resistant Acinetobacter infection and their susceptibility patterns in a tertiary care hospital. Niger Med J. 2012;53:126-8.
- [Google Scholar]
- The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42:692-9.
- [Google Scholar]
- The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol. 2012;3:148.
- [Google Scholar]
- Comparison between bacteremia caused by carbapenem resistant Acinetobacter baumannii and Acinetobacter nosocomialis. BMC Infect Dis. 2013;13:311.
- [Google Scholar]
- Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282.
- [Google Scholar]
- Effect of desiccation on the ultrastructural appearances of Acinetobacter baumannii and Acinetobacter lwoffii. J Clin Pathol. 1998;51:786-8.
- [Google Scholar]
- Resistance to disinfectants in epidemiologically defined clinical isolates of Acinetobacter baumannii. J Hosp Infect. 2007;66:174-81.
- [Google Scholar]





