Translate this page into:
Molecular epidemiology of Carbapenemase-encoding genes and comparative evaluation of carbapenem MIC with genotypic carbapenem resistance in Klebsiella isolates from neonatal sepsis cases
*Corresponding author: Nisha Goyal, Department of Microbiology, University College of Medical Sciences and Guru Teg Bahadur Hospital, Delhi, India. drnishagoyalucms@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Gangar S, Singh NP, Goyal N, Mohapatra S, Das S, Batra P. Molecular epidemiology of Carbapenemase-encoding genes and comparative evaluation of carbapenem MIC with genotypic carbapenem resistance in Klebsiella isolates from neonatal sepsis cases. J Lab Physicians. 2024;16:200-6. doi: 10.25259/JLP_23_2023
Abstract
Objectives:
The objective of this study was to determine the molecular epidemiology of Carbapenemase-encoding genes in Klebsiella isolates from neonatal sepsis cases and comparative evaluation of carbapenem minimum inhibitory concentration (MIC) with genotypic carbapenem resistance.
Materials and Methods:
One hundred cases of neonatal sepsis with blood cultures positive for Klebsiella spp. were included in the study. MIC for imipenem and meropenem was determined by Epsilometer-test. Antimicrobial susceptibility testing (AST) was performed by modified Kirby Bauer disc diffusion method. All the isolates of Klebsiella spp. were tested for the presence of beta-lactamase Klebsiella pneumoniae carbapenemase (blaKPC ), beta-lactamase New Delhi metalloβ-lactamase-1(blaNDM-1), beta-lactamase imipenemase (blaIMP), beta-lactamase Verona imipenemas e (blaVIM) genes by multiplex polymerase chain reaction (PCR) and uniplex PCR for beta-lactamase oxacillinase-48 (blaOXA-48). Comparison of individual antibiotic susceptibility between carbapenemase-encoding gene positive and negative Klebsiella spp. isolates was performed.
Statistical analysis:
Statistical analysis was done using the Fisher’s exact test. P < 0.05 was considered significant.
Results:
The prevalence of carbapenemase-encoding genes in Klebsiella spp. was 16%. Most predominant carbapenemase-encoding gene was blaOXA-48 gene (12%) followed by blaNDM-1 gene (6%). Coexpression of both blaOXA-48 and blaNDM-1 was observed in 2% of isolates. All the Klebsiella spp. isolates harboring the carbapenemases gene (100%) had resistant MIC values for Meropenem, whereas, for imipenem, only 75% of isolates had resistant MIC values.
Conclusions:
Determination of prevalence of carbapenemase-encoding genes is of paramount importance in the development of effective antibiotic policies at various levels.
Keywords
Carbapenemase-encoding genes
Carbapenem-resistant Enterobacteriales
OXA-48
New Delhi metallo-b-lactamase
Coexpression OXA-48 and New Delhi metallo-b-lactamase
Klebsiella pneumoniae
INTRODUCTION
The past two decades have witnessed an increase in the incidence and spread of multidrug-resistant organisms like Carbapenem-resistant Enterobacteriales (CRE). CRE poses a considerable threat to mankind as they are resistant to almost all the available beta-lactam antimicrobials,including one or more carbapenem drugs along with fluoroquinolones, and aminoglycosides. The treatment options are either very few or non-existent for this group of multidrug-resistant bacteria.[1]
Klebsiella pneumoniae (K. pneumoniae) is the most notorious member of CRE group that is frequently associated with life-threatening infections. This pathogen can be resistant to the repertoire of available antibiotics. The isolation of K. pneumoniae in increased frequencies from hospital environments,including intensive care units especially in developing countries is alarming.[2-4] K. pneumoniae is very frequently associated with oxacillinases (OXA)-48 like and New Delhi metallo-b-lactamase (NDM) carbapenemase-encoding genes globally. Acquisition and transmission of potent transferable plasmid-encoded carbapenemases such as Klebsiella pneumoniae Carbapenemase (KPC), NDM, Imipenemase (IMP), Verona integron-encoded metallo-beta-lactamase (VIM), and oxacillinase-48 like enzymes is common in multi-drug resistant and hypervirulent strains of Klebsiella spp.[3]
India is endemic to CRE harboring carbapenemase-encoding genes of OXA-48,such as OXA-181 and OXA-232 and NDM-1 variants, which were first described in K. pneumonia.[5,6] The sporadic outbreaks of various members of CRE possessing other carbapenemase-encoding genes such as IMP, KPC, and VIM have also been reported over recent years.[7,8] Noticeably, the circulating carbapenemase strains of pathogenic bacteria may vary geographically or even at different locations within the same healthcare setting. Phenotypic determination of potential carbapenem resistance due to the presence of carbapenemase-encoding gene and its impact on the susceptibility to other antimicrobial agents varies widely according to the individual carbapenemase-encoding gene present.[1] OXA-48 like carbapenemases is known to possess weak carbapenem hydrolytic activity and, thus, is likely to be missed by phenotypic detection methods.[9,10] Therefore, the molecular identification of resistance mechanisms in circulating strains of CRE pathogens is crucial for healthcare settings to effectively manage life-threatening infections without unwarranted delay. Furthermore, the availability of information regarding the prevalent resistant strains can play an important role in containment of these infections.
Neonatal sepsis is the third leading cause of neonatal mortality. As per United Nations Children’s Fund, the neonatal mortality rate in India was 21.7/1000 live births in the year 2022.[11] Carbapenem-resistant K. pneumoniae is often known for frequently causing outbreaks in neonatal intensive care units (NICUs), thus warrants its molecular characterization pertaining to the presence of carbapenemase-encoding genes.[12] Often the inadequacy of molecular diagnosis of CRE and their underlying resistance mechanisms in resource-limited settings contribute to the inadvertent spread of these resistant isolates. The growing complexities surrounding the treatment of life-threatening infections caused by CRE demands an understanding of its local epidemiology for providing adequate empirical coverage and antimicrobial resistance control initiatives.
Numerous studies have addressed the dissemination of CRE, but the data on neonatal sepsis are limited. Earlier studies reported the presence of a single carbapenemase gene in K. pneumoniae, but recent years have shown the emergence of coproduction of multiple carbapenemases.[13,14] Furthermore, NDM producing K. pneumoniae was more prevalent previously in neonatal infections. Besides, recent years have noted a significant emergence of highly transferable OXA-48 class D carbapenemase in Enterobacteriales.[15,16] Therefore, the present study attempted to determine the molecular epidemiology of Carbapenemase-encoding genes and comparative evaluation of carbapenem minimum inhibitory concentration (MIC) with genotypic carbapenem resistance in Klebsiella isolates from neonatal sepsis cases.
MATERIALS AND METHODS
A prospective study was conducted by the molecular laboratory of the Department of Microbiology in collaboration with NICU and the Department of Pediatrics. Post obtaining the approval from the Institutional Ethics Committee-Human Research, the study was extended for over a period of one year, starting July 2019 to June 2020. The first 100 neonates admitted to NICU with clinical signs and symptoms suggestive of neonatal sepsis who had blood cultures positive for Klebsiella spp. were included in the study. Sepsis screening was performed in the clinically suspected cases. The criteria included total leukocyte count (<500/mm3), absolute neutrophil count (<1800/mm3), immature to a total neutrophil ratio (>20%), C-reactive protein (>1 mg/dL), and micro erythrocyte sedimentation rate (15 mm or more in the 1st h). Sepsis screening was considered positive if any two of these parameters were positive.[17]
All these non-duplicate Klebsiella spp. isolates from blood samples of neonatal sepsis patients were identified and processed further using the standard laboratory guidelines. Antimicrobial susceptibility testing (AST) was performed by modified Kirby Bauer disc diffusion method using the amikacin (30 μg), aztreonam (30 μg), azithromycin (15 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefixime (5 μg), ciprofloxacin (5 μg), cotrimoxazole (25 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), and piperacillin-tazobactam (100/10 μg) antibiotic discs (HiMedia laboratories, India).
Minimum Inhibitory Concentration (MIC for imipenem and meropenem was determined by Epsilometer-test using Imipenem Ezy MIC™ Strip (IPM) (0.002–32 μg/mL) and Meropenem Ezy MIC™ Strip (MRP) (0.002–32 μg/mL) (HiMedia Laboratories, India). K. pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC BAA-1706 were used as positive and negative controls. For both the antibiotics, imipenem and meropenem, isolates were considered susceptible (MIC ≤1 μg/mL, intermediate (MIC of 2 μg/mL), and resistant (MIC ≥4 μg/mL) after incubation of 18 h at 35–37°C. The AST and interpretation of results were performed as per Clinical and Laboratory Standards Institute guidelines.[18]
The deoxyribonucleic acid (DNA) was extracted (HIPURATM bacterial genomic DNA purification kit from HiMedia laboratories, India) by spin-column procedure. All the isolates of Klebsiella spp. were tested for the presence of beta-lactamase Klebsiella pneumoniae carbapenemase (blaKPC), beta-lactamase New Delhi metallo-β- lactamases-1(blaNDM-1), beta-lactamase imipenemase (blaIMP), beta-lactamase Verona imipenemase (blaVIM) genes by multiplex polymerase chain reaction (PCR) and uniplex PCR for beta-lactamase oxacillinase-48 (blaOXA-48) using the primer sequences for target genes mentioned in Table 1.[19,20]
| S. No. | GENES | FP/RP | PRIMER SEQUENCE ( 5’-3’) | SIZE (bp) |
|---|---|---|---|---|
| 1. | NDM-1 | FP | GCATAAGTCGCAATCCCCG | 237 |
| RP | CTTCCTATCTCGACATGCCG | |||
| 2. | VIM | FP | GTTTGGTCGCATATCGCAAC | 382 |
| RP | AATGCGCAGCACCAGGATAG | |||
| 3. | IMP | FP | GAAGGCGTTTATGTTCATAC | 578 |
| RP | GTAAGTTTCAAGAGTGATGC | |||
| 4. | KPC | FP | TCGAACAGGACTTTGGCG | 201 |
| RP | GGAACCAGCGCATTTTTGC | |||
| 5. | OXA-48 | FP | GCTTGATCGCCCTCGATT | 281 |
| RP | GATTTGCTCCGTGGCCGAAA |
NDM-1: New Delhi metallo-β-lactamase-1, VIM: Verona integron-encoded metallo-beta-lactamase, IMP: Imipenemase, KPC: Klebsiella pneumoniaeCarbapenemase, OXA-48-like: Oxacillinases, FP: Forward Primer, RP: Reverse Primer, bp: Base pair
Cycling conditions for blaKPC, blaNDM-1, blaIMP, and blaVIM genes Multiplex PCR: [19] For amplification, 30 cycles were done at 94°C for 1 min, 54°C for 1 min, and 72°C for one and a half minutes followed by a final extension step of 5 min at 72°C.
Cycling conditions for blaOXA-48 gene Uniplex PCR:[20] For amplification, initial denaturation at 94°C for 10 min; 30 cycles of 94°C for 40 s, 60°C for 40 s, and 72°C for 1 min followed by a final elongation step at 72°C for 7 min. The annealing temperature was optimal at 57°C.
The amplified products were then run on 1.5% Agarose gel (HiMedia laboratories India) at 100 Volts for 45 min to 1 h and viewed under UV light (G: BOX, SynGene). The gel was then photographed using GeneSnap from SynGene [Figure 1].
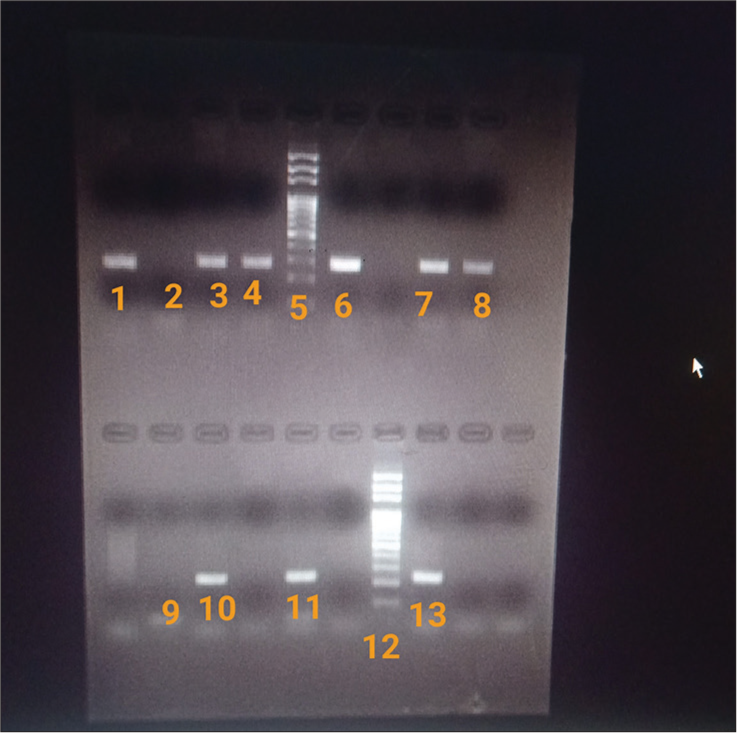
- Gel electrophoresis showing bands of DNA and 100bp DNA ladder in GeneSnap software. 1: Positive control for blaNDM-1 gene; 2,9: Negative control (Distilled water); 5, 12: DNA ladder; 3,4,6,7,8: Test (positive for blaNDM-1 gene); 10: Positive control for blaOXA-48 gene; 11,13: Test (positive for blaOXA-48 gene). blaNDM-1: beta-lactamase New Delhi metallo-β-lactamases-1, blaOXA-48: Oxacillinases.
Statistical analysis
Comparison of individual antibiotic susceptibility between carbapenemase-encoding gene positive and negative Klebsiella spp. isolates was performed using the Fisher’s exact test. P-values were calculated for the analysis of the difference in antibiotic susceptibility between carbapenemase-encoding gene positive and negative Klebsiella spp. Isolates. Relevant frequencies and percentages were calculated.
RESULTS
Distribution of carbapenemase-encoding genes among Klebsiellas spp. isolates
The overall prevalence of carbapenemase-encoding genes in Klebsiella spp. isolates from blood samples of neonatal sepsis cases admitted in NICU was 16% (16/100). The distribution of various carbapenemase-encoding genes is shown in Table 2 and Figure 2. The most predominant carbapenemase-encoding gene was blaOXA-48 gene (12%), followed by blaNDM-1 gene (6%). None of the isolates harbored carbapenemase-encoding genes of blaIMP, blaVIM, and blaKPC. The coexpression of both blaOXA-48 and blaNDM-1 was observed in 2% of isolates.
| Carbapenemase-encoding gene/s | Present (%) | Absent (%) |
|---|---|---|
| KPC | 0 | 100 |
| NDM-1 | 6 | 94 |
| IMP | 0 | 100 |
| VIM | 0 | 100 |
| OXA-48 | 12 | 88 |
| Coexpression of blaOXA-48 and blaNDM-1 | 2 | 98 |
| Total | 16 | 82 |
KPC: Klebsiella pneumoniaeCarbapenemase, NDM: New Delhi metallo-β-lactamase, IMP: Imipenemase, VIM: Verona integron-encoded metallo-beta-lactamase, OXA-48-like: Oxacillinases, NICU: Neonatal intensive care unit. n: Total number of tested isolates.

- Prevalence of carbapenemase-encoding genes in Klebsiella spp. isolates from blood samples of neonatal sepsis cases admitted in neonatal intensive care unit (n = 100). n: Specify total number of tested isolates, KPC: Klebsiella pneumoniae Carbapenemase, NDM-1: New Delhi metallo-β-lactamase-1, IMP: Imipenemase, VIM: Verona integron-encoded metallo-beta-lactamase, OXA-48-like: Oxacillinases.
Comparative distribution of carbapenemase-encoding genes in different MIC cutoff categories of imipenem and meropenem
Comparative distribution of carbapenemase-encoding genes in different MIC cutoff categories of imipenem and meropenem among Klebsiellas spp. isolates from blood samples of neonatal sepsis cases admitted in NICU are shown in Table 3. All the Klebsiella spp. isolates harboring the carbapenemases gene (100%) had resistant MIC values for meropenem (≥4 μg/mL), whereas, for imipenem, only 75% of isolates had resistant MIC values (≥4 μg/mL). For imipenem, 18.7% of carbapenemase-encoding genes harboring Klebsiella spp. had susceptible (≤1 μg/mL) and 6.25% had intermediate MIC values. Out of three isolates with susceptible MIC values for imipenem, two were harboring blaOXA-48 (MIC 0.19 μg/mL and 0.25 μg/mL),and one had blaNDM-1 gene (MIC 0.75 μg/mL). The isolate with intermediate MIC harbored blaOXA-48 gene (MIC 2 μg/mL). A significant association was observed between the presence of carbapenemase-encoding genes and MIC values for imipenem and meropenem (P < 0.001 and P = 0.016, respectively).
| Carbapenemase Genes (N=100) | IMIPENEM n(%) | MEROPENEM n(%) | ||||
|---|---|---|---|---|---|---|
| Susceptible (≤1 μg/mL) | Intermediate (2 μg/mL) | Resistant (≥4 μg/mL) | Susceptible (≤1 μg/mL) | Intermediate (2 μg/mL) | Resistant (≥4 μg/mL) | |
| Present (n=16) | 3 (18.7) | 1 (6.25) | 12 (75) | 0 | 0 | 16 (100) |
| Absent (n=84) | 56 (66.6) | 16 (19) | 12 (14.3) | 40 (47.6) | 17 (20.2) | 27 (32.1) |
| Total (n=100) | 59 (59) | 17 (17) | 24 (24) | 40 (40) | 17 (17) | 43 (43) |
MIC: Minimum inhibitory concentration, NICU: Neonatal intensive care unit, n: Total number of tested isolates
Comparative distribution of susceptibility to various antimicrobial agents in Klebsiella spp. isolates with and without genotypic carbapenem resistance
Table 4 shows the comparative distribution of susceptibility to various antimicrobial agents in Klebsiellas spp. isolates with and without genotypic carbapenem resistance. Resistance to amikacin, aztreonam, ceftazidime, cefotaxime, cefixime, ciprofloxacin, cotrimoxazole, gentamicin, imipenem, meropenem, and piperacillin-tazobactam were higher among Klebsiella spp. isolates harboring the carbapenemases gene. The difference in the susceptibility among two groups of Klebsiellas spp. isolates with genotypic carbapenem resistance and Klebsiellas spp. isolates without genotypic carbapenem resistance were statistically significant for all the tested antibiotics (P < 0.05) except amikacin (P > 0.05).
| Antimicrobial (μg/disc) | PCR Positive isolates (n=16) | PCR Negative isolates (n=84) | P-value | ||
|---|---|---|---|---|---|
| Sensitive n(%) | Resistance n(%) | Sensitive n(%) | Resistance n(%) | ||
| Amikacin (30) | 6 (37.5) | 10 (62.5) | 53 (63) | 31 (36.9) | 0.093 |
| Aztreonam (30) | 0 (0) | 16 (100) | 44 (52.3) | 39 (46.4) | 0.0001 |
| Ceftazidime (30) | 1 (6.25) | 15 (93.75) | 31 (36.9) | 53 (63) | 0.018 |
| Cefotaxime (30) | 1 (6.25) | 15 (93.75) | 32 (38) | 52 (61.9) | 0.0178 |
| Cefixime (5) | 1 (6.25) | 15 (93.75) | 33 (39.2) | 51 (60.7) | 0.009 |
| Ciprofloxacin (5) | 1 (6.25) | 15 (93.75) | 39 (46.4) | 45 (53.5) | 0.002 |
| Cotrimoxazole (25) | 1 (6.25) | 15 (93.75) | 44 (52.3) | 40 (47.6) | 0.0006 |
| Gentamicin (10) | 2 (12.5) | 14 (87.5) | 64 (76.1) | 20 (23.8) | 0.0001 |
| Imipenem (10)* | 5 (31.25) | 9 (56.25) | 76 (90.4) | 8 (9.5) | 0.0001 |
| Meropenem (10)^ | 1 (6.25) | 14 (87.5) | 63 (75) | 19 (22.6) | 0.0001 |
| Piperacillin-Tazobactam (100/10)** | 3 (18.75) | 13 (81.25) | 59 (70.2) | 23 (27.3) | 0.0001 |
PCR: Polymerase chain reaction. *Imipenem – Out of 16 PCR positive Klebsiella isolates, 2 (12.5%) isolates had intermediate zone diameters. ^Meropenem – Out of 16 PCR positive Klebsiella isolates, 1 (6.25%) isolate had an intermediate zone diameter, and among 84 PCR negative isolates, 2 (2.4%) had an intermediate zone diameter for meropenem. **Piperacillin-Tazobactam – 2 (2.4%) PCR negative Klebsiella Isolates had intermediate zone diameter. The P-value ≤0.05 was considered statistically significant”
DISCUSSION
Neonatal sepsis remains the third leading cause of neonatal mortality. The bacteriological profile responsible for neonatal sepsis varies geographically. The most frequent offenders of early-onset sepsis and late-onset sepsis in developing nations like India are K. pneumoniae and Staphylococcus aureus.[21] The production of carbapenemases by Enterobacterales isolates especially Klebsiella spp., associated with invasive infections, gut colonization, or hospital-acquired infections, has been detected across the globe.[22,23] These superbugs causing infections in vulnerable newborns are present everywhere, including the birth canal, incubators, hands of healthcare workers, and even the medical equipment like nasogastric tubes. Premature or low-birth-weight infants needing extended hospital stay or dependency on life support systems have significantly elevated risk of infection by these pathogens. The carbapenemase producers play a significant role in triggering outbreaks by transferring plasmid-encoded carbapenemase genes, thus posing an exceptionally difficult challenge in the containment of these infections.[24] Many Indian studies have documented the high prevalence of bloodstream infections caused by multidrug resistant Klebsiella spp. isolates in high-risk settings of NICUs.[25,26]
The predominant mechanism of resistance in the present study was the presence of blaOXA-48 (12%) followed by blaNDM-1 (6%). The coexpression of both blaOXA-48 and blaNDM-1 was observed in 2% of isolates. However, another study from India has reported blaOXA48-like expression in 13%, blaNDM in 19%, and coexpression of blaNDM and blaOXA48-like in 28% isolates bloodstream isolates of K. pneumonia.[27] In line with our findings, this research has also reported the predominant carbapenem-encoding genes as blaNDM and blaOXA48. Laolerd et al.,[28] have reported the presence of at least one of the carbapenemase genes in 77.7% of CRE isolates (predominantly K. pneumoniae). The same study also documented that the prevalence of blaNDM (71.75%) and blaOXA48-like (50.22%) genes among CRE isolates and coexpression of both blaNDM and blaOXA48-like genes was 32.73% among K. pneumoniae isolates.[28] The differences in the distribution of blaNDM , blaOXA48, and their coexpression can be attributed to the geographical variations.
The present study documented blaOXA-48 carbapenem-encoding gene to be most prevalent in a tertiary care high-volume center of north India. The OXA-48-like enzymes are hidden threats as they possess a low level of carbapenem hydrolytic activity. This mechanism of resistance remains underreported by standard laboratory procedures and subsequent treatment failures.[10] In the present study, intermediate and susceptible MIC values were also observed along with resistant MIC values for imipenem in blaOXA-48 harboring isolates. This could be probably due to the absence of coresistance mechanisms in OXA-48 like producers. In the absence of other resistance mechanisms, OXA-48, like producers,can show lower or intermediate MIC values for carbapenems. Thus, their true prevalence rates are hard to estimate.[29,30] In our study, all the carbapenemase gene-harboring isolates had resistant MIC values for meropenem. Thus, meropenem is a better predictor of carbapenemase production in comparison to imipenem.
The frequent use of carbapenems and other beta-lactam antibiotics has led to increased resistance in the bacteria,and the rapid fluctuation in their susceptibility pattern challenges the treatment options for serious infections. Current guidelines from the World Health Organization recommend the use of ampicillin, extended-spectrum cephalosporins (cefotaxime), and aminoglycosides (gentamicin and amikacin) for the treatment of neonatal sepsis. This protocol is widely followed in various healthcare settings.[31] However, the production of AmpC beta-lactamases and extended-spectrum beta-lactamases has led to increasing dependency on carbapenems as next line of treatment. Among carbapenems, meropenem is the preferred carbapenem in neonates as the safety profiles of other carbapenems are not well documented in this patient group.[32]
The OXA-48 producers hydrolyze penicillins with greater efficiency and carbapenems at a lower extent, sparing expanded-spectrum cephalosporins, whereas MBLs (NDM-1) are more potent, they can hydrolyze almost all available beta-lactam antibiotics, including carbapenems along with extended-spectrum cephalosporins, aminoglycosides, aztreonam, and fluoroquinolones.[33] Optimizing treatment strategies for these infections become a challenge due to the lack of knowledge about the circulating carbapenemases. We compared the distribution of susceptibility to routine antimicrobial agents in isolates with and without genotypic carbapenem resistance. Despite the inability of OXA-48 producers to hydrolyze expanded-spectrum cephalosporins, high resistance rates (94%) to these cephalosporins were observed in our study. This additional resistance to expanded-spectrum cephalosporins among OXA-48 producers can be due to the presence of other mechanisms of resistance including coexpression of both OXA-48 and NDM-1. Imipenem was the most effective antimicrobial agent in the absence of any carbapenemase-encoding gene and amikacin was most effective in the presence of carbapenemase-encoding genes followed by imipenem. Similar results showing carbapenem drugs being most effective against Klebsiella were also reported in study by Mohsen et al.[34]
Considering the widespread prevalence of multidrug-resistant Klebsiella spp. as a causative agent of life-threatening infections, it is crucial to understand the mechanisms of resistance behind the last resort drugs of carbapenems. Furthermore, their therapeutic utility can be largely restricted by the presence of certain carbapenemase-encoding genes.[35] Thus, the determination of carbapenem resistance by phenotypic methods should be supplemented with genotypic assessment. The knowledge of prevailing strains of carbapenemases can aid in the implementation of antibiotic cycling and formulation of effective hospital antibiotic policies. This has the potential to significantly impact patient care in terms of reduced morbidity, mortality, and duration of hospital stay. However, further elaborative studies with larger sample sizes and diverse clinical conditions are warranted.
Carbapenems are broad-spectrum higher-level antibiotics and are often considered as last resort antibiotics. The emergence of carbapenemases is a major healthcare problem. The susceptibility of carbapenem-producing CRE to carbapenem drugs may vary depending on the molecular class of carbapenemase produced. The distribution of circulating strains is not uniform and varies geographically. Therefore, the determination of prevalence of carbapenemase-encoding genes may be of paramount importance in the development of effective antibiotic policies at various levels.
CONCLUSIONS
Early detection and knowledge of the prevalence of carbapenemase encoding genes significantly benefit patient care by lowering morbidity, mortality, and hospitalization duration. This also impacts the formulation of combination antibiotic therapies, and hospital antibiotic policies and forms the basis of antibiotic cycling as an important component of infection control in NICU.
Ethical approval
The research/study approved by the Institutional Review Board at UCMS-Institutional Ethics Committee-Human Research (IEC-HR), number IEC-HR/2018/36/73R, dated 26/10/2018.
Declaration of patient consent
Patient’s consent is not required as patients identity is not disclosed or compromised.
Conflict of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- The problem of carbapenemase-producing-carbapenem-resistant Enterobacteriaceae detection. J Clin Microbiol. 2016;54:529-34.
- [CrossRef] [PubMed] [Google Scholar]
- An overview of carbapenem-resistant Klebsiella pneumoniae. Rev Med Microbiol. 2014;25:7-14.
- [CrossRef] [Google Scholar]
- Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front Med. 2018;5:225.
- [CrossRef] [PubMed] [Google Scholar]
- Klebsiella pneumoniae as a key tracker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131-9.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of antibiotic resistance determinants and their transmissibility among clinically isolated carbapenem-resistant Escherichia coli from South India. Med Princ Pract. 2018;27:428-35.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of carbapenemase-mediated resistance among Enterobacteriaceae bloodstream isolates: A molecular study from India. Indian J Med Microbiol. 2017;35:421-5.
- [CrossRef] [PubMed] [Google Scholar]
- Structure, genetics and worldwide spread of New Delhi metallo-b-lactamase (NDM): A threat to public health. BMC Microbiol. 2017;17:101.
- [CrossRef] [PubMed] [Google Scholar]
- The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J Infect Dis. 2017;215:S28-36.
- [CrossRef] [PubMed] [Google Scholar]
- Phenotypic and genotypic characterization of Enterobacteriaceae producing oxacillinase-48-like carbapenemases, United States. Emerg Infect Dis. 2018;24:700-9.
- [CrossRef] [PubMed] [Google Scholar]
- Treatment of infections by OXA-48-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2018;62:e01195-18.
- [CrossRef] [PubMed] [Google Scholar]
- Neonatal mortality. 2023. UNICEF data. Available from: https://data.unicef.org/topic/child-survival/neonatal-mortality [Last accessed on 2023 Aug 22]
- [Google Scholar]
- KPC with ESBL: A multistarrer tragedy. Ann Trop Med Public Health. 2016;9:16-8.
- [CrossRef] [Google Scholar]
- Extensively-drug resistant Klebsiella pneumoniae recovered from neonatal sepsis cases from a major NICU in Egypt. Front Microbiol. 2020;11:1375.
- [CrossRef] [PubMed] [Google Scholar]
- OXA-181-like carbapenemases in Klebsiella pneumoniae ST14, ST15, ST23, ST48, and ST231 from septicemic neonates: coexistence with ndm-5, resistome, transmissibility, and genome diversity. mSphere. 2021;6:e01156-20.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of co-transfer of plasmid-mediated fluoroquinolone resistance genes and blaNDM gene in Enterobacteriaceae causing neonatal septicaemia. Antimicrob Resist Infect Control. 2019;8:46.
- [CrossRef] [PubMed] [Google Scholar]
- Spread and exchange of bla NDM-1 in hospitalized neonates: Role of mobilizable genetic elements. Eur J Clin Microbiol Infect Dis. 2017;36:255-65.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical profile and outcome of acute kidney injury in neonatal sepsis in a tertiary care centre (Doctoral dissertation, Rajiv Gandhi University of Health Sciences India) 2016
- [CrossRef] [Google Scholar]
- Performance standards for antimicrobial susceptibility testing; Twenty-seventh informational supplement. In: CLSI document M100-S29. Wayne, PA: Clinical and Laboratory Standards Institute; 2019.
- [Google Scholar]
- Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among gram negative bacilli. J Clin Diagn Res. 2014;8:DC23-6.
- [CrossRef] [PubMed] [Google Scholar]
- Development of a set of multiplex PCR assays for the detection of genes encoding important b-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490-5.
- [CrossRef] [PubMed] [Google Scholar]
- Neonatal sepsis: Mortality and morbidity in neonatal sepsis due to multidrug-resistant (MDR) organisms: Part 1. Indian J Pediatr. 2020;87:117-21.
- [CrossRef] [Google Scholar]
- Rapid detection of KPC, NDM, and OXA-48-like carbapenemases by real-time PCR from rectal swab surveillance samples. J Clin Microbiol. 2015;53:2731-3.
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenem resistant Enterobacteriaceae neonatal gut colonization: A future concern in healthcare settings. Indian J Microbiol Res. 2018;5:348-54.
- [CrossRef] [Google Scholar]
- Neonatal sepsis: The impact of carbapenem-resistant and hypervirulent Klebsiella pneumoniae. Front Med. 2021;8:634349.
- [CrossRef] [PubMed] [Google Scholar]
- Outbreak of carbapenemase-producing Klebsiella pneumoniae blood stream infections in neonatal intensive care unit. Int J Curr Microbiol Appl Sci. 2016;5:727-33.
- [CrossRef] [Google Scholar]
- Prevalence of neonatal septicaemia in a tertiary care hospital in Mandya, Karnataka, India. Int J Res Med Sci. 2016;4:2812-6.
- [CrossRef] [Google Scholar]
- Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog Glob Health. 2017;111:240-6.
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenemase-producing carbapenem-resistant Enterobacteriaceae from Bangkok, Thailand, and their detection by the Carba NP and modified carbapenem inactivation method tests. Microb Drug Resist. 2018;24:1006-11.
- [CrossRef] [PubMed] [Google Scholar]
- OXA-48-like carbapenemases: The phantom menace. J Antimicrob Chemother. 2012;67:1597-606.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D b-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41:325-9.
- [CrossRef] [PubMed] [Google Scholar]
- Serious neonatal bacterial infections caused by Enterobacteriaceae (including septicemia and meningitis) In: Kimberlin DW, Barnett ED, Lynfield R, Sawyer MH, eds. Red book: 2021-2024 Report of the committee on infectious diseases (32nd ed). United States: American Academy of Pediatrics; 2021. p. :311.
- [CrossRef] [Google Scholar]
- Tables of antibacterial drug dosages In: Kimberlin DW, Barnett ED, Lynfield R, Sawyer MH, eds. Red book: 2021-2024 report of the committee on infectious diseases (32nd ed). United States: American Academy of Pediatrics; 2021. p. :876.
- [CrossRef] [Google Scholar]
- Molecular mechanisms, epidemiology, and clinical importance of b-lactam resistance in Enterobacteriaceae. Int J Mol Sci. 2020;21:5090.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial susceptibility of Klebsiella pneumoniae and Escherichia coli with extended-spectrum b-lactamase associated genes in hospital Tengku Ampuan Afzan, Kuantan, Pahang. Malays J Med Sci. 2016;23:14-20.
- [Google Scholar]
- Diversity of resistance mechanisms in carbapenem-resistant Enterobacteriaceae at a health care system in Northern California, from 2013 to 2016. Diagn Microbiol Infect Dis. 2019;93:250-7.
- [CrossRef] [PubMed] [Google Scholar]






