Translate this page into:
Phenotypic Detection of Carbapenemase Production in Carbapenem-Resistant Enterobacteriaceae by Modified Hodge Test and Modified Strip Carba NP Test
Address for correspondence: Babita Sharma, MD, 31 Shree Ganesh Colony Mahesh Nagar, 80 Feet Road, Jaipur 302019, Rajasthan, India (e-mail: babitasharma13@yahoo.com). Nisha Patidar, MD, B-7; L-1126, Vrinda Gardens, Jagatpura, Jaipur 302017, Rajasthan, India (e-mail: sweetdr.nisha@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
Carbapenems are last resort antibiotics for multidrug-resistant Enterobacteriaceae. However, resistance to carbapenem is increasing at an alarming rate worldwide leading to major therapeutic failures and increased mortality rate. Early and effective detection of carbapenemase producing carbapenem-resistant Enterobacteriaceae (CRE) is therefore key to control dissemination of carbapenem resistance in nosocomial as well as community-acquired infection. The aim of present study was to evaluate efficacy of Modified strip Carba NP (CNP) test against Modified Hodge test (MHT) for early detection of carbapenemase producing Enterobacteriaceae (CPE).
Material and Methods
Enterobacteriaceae isolated from various clinical samples were screened for carbapenem resistance. A total of 107 CRE were subjected to MHT and Modified strip CNP test for the detection of CPE.
Statistical Analysis
It was done on Statistical Package for the Social Sciences (SPSS) software, IBM India; version V26. Nonparametric test chi-square and Z-test were used to analyze the results within a 95% level of confidence.
Results
Out of 107 CRE, 94 (88%) were phenotypically confirmed as carbapenemase producer by Modified strip CNP test and 46 (43%) were confirmed by Modified Hodge Test (MHT). Thirty-eight (36%) isolates showed carbapenemase production by both MHT and CNP test, 56 isolates (52%) were CNP test positive but MHT negative, eight (7%) isolates were MHT positive but CNP test negative and five (5%) isolates were both MHT and CNP test negative. There is statistically significant difference in efficiency of Modified CNP test and MHT (p < 0.05).
Conclusion
Modified strip CNP test is simple and inexpensive test which is easy to perform and interpret and gives rapid results in less than 5 minutes. It has high degree of sensitivity and specificity. Modified strip CNP test shows significantly higher detection capacity for carbapenemase producers as compared with MHT.
Keywords
carbapenem-resistant Enterobacteriaceae
carbapenemase producing Enterobacteriaceae
modified Hodge test
modified strip Carba NP test
Introduction
Members of Enterobacteriaceae family are gram negative, rod-shaped facultative anaerobes mainly colonizing the intestinal tracts of humans and animals and are a common cause of community-associated as well as health care-associated infections.[1]
In the past, most of the first line and low-cost antimicrobial drugs such as penicillin and first and second generation cephalosporins have been used effectively for the management of gram-negative bacterial infections. However, drug-resistant organisms acquire resistance to these first-line antibiotics, thereby necessitating the need of second-line drugs like the third and fourth generation cephalosporins. Further emergence of β lactamases, extended spectrum β lactamases (ESBL),[2] and AmpC β lactamases[3] producing bacterial strains lead to the development of multidrug-resistant (MDR) Enterobacteriaceae.
Carbapenems such as ertapenem, imipenem, meropenem, and doripenem have proven efficacy in severe infections caused by these MDR Enterobacteriaceae; therefore, they are frequently used as last resort therapeutic options. Carbapenems have the widest spectrum of antibacterial activities[4] and are also active against the chromosomal cephalosporinases and ESBL, therefore are preferred antibiotics in case of invasive or life-threatening infections. However, in recent years, carbapenem-resistant Enterobacteriaceae (CRE) has emerged in the community as a major health threat.
Carbapenem resistance was first reported sporadically in the mid-1990s from the United States and since then reports of carbapenem resistance outbreaks are increasing worldwide at an alarming rate.[5] In addition to carbapenem resistance, CRE often carry genes that confer high levels of resistance to many other antimicrobials, therefore these bacteria are difficult to treat and are associated with high mortality.[6]. The mechanisms of development of carbapenem resistance in Enterobacteriaceae are complex because they involve a broad range of organisms and are mediated by different mechanisms. Broadly, CRE can be carbapenemase producing CRE (CP-CRE) or noncarbapenemase producing CRE (non-CP-CRE).[7]
CP-CRE also known as carbapenemase producing Enterobacteriaceae (CPE) encompasses organisms that are resistant to carbapenems due to the production of a carbapenemase enzyme. Carbapenemases are specific β-lactamases enzymes produced by gram-negative bacteria having ability to hydrolyze carbapenems and other β-lactam antibiotics like penicillin and cephalosporins. Various carbapenemases have been reported in Enterobacteriaceae which can be classified as Class A: plasmid encoded Klebsiella pneumoniae carbapenemase (KPC) and Guiana extended spectrum variant,[8] Class B metallo-β-lactamase (MBL) including imipenemases, Verona integron-encoded metallo-β-lactamase (VIM), New Delhi metallo-β-lactamases (NDM),[8-10] Class D Oxacillinase (OXA) comprises OXA-48 derivatives.[11] India is endemic for NDM type MBL while KPC producing isolates are sporadic.[12-14]
Noncarbapenemase producing carbapenem-resistant Enterobacteriaceae (non CP-CRE) encompasses organisms that are resistant to carbapenems due to mechanisms other than carbapenemase production like increase in bacterial outer membrane permeability, augmenting drug efflux, mutations or loss of outer membrane porins preventing entry of antibiotics, or due to overexpression of β-lactamases like ESBL and AmpC β-lactamases.[7,15]
Noncarbapenemase-related mechanism of carbapenem resistance are nontransferable while carbapenemase production is plasmid encoded. Thus, CPE have potential for widespread transmission, resulting in increased frequency of carbapenem resistance on a worldwide basis.[8]
Currently, CRE are a problem not confined to individual facilities but are affecting entire communities and nations. High incidence of treatment failures, morbidity, and mortality in many infectious diseases are strongly correlated with carbapenem resistance. There are limited antimicrobials effective against carbapenemase-producing bacteria. Most hospitals have limited facilities and resources to properly confirm the presence of carbapenemase production in clinical isolates. This is the need of hour to have a laboratory test which is easy to perform, cost effective, and at the same time is highly sensitive and specific. Genotypic methods like real time polymerase chain reaction-restricted fragment length polymorphism (PCR-RFLP)[16,17] and whole genome sequencing, and other techniques like matrix assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS)[18,19] have overall good sensitivities and specificities but require trained microbiologists, expensive equipment, and are time consuming. In addition, it may fail to detect unknown novel carbapenemase genes.[20] On the other hand, phenotypic tests like Modified Hodge test (MHT) and Carba NP (CNP) test are inexpensive, easier to perform, and require minimal training. MHT is highly sensitive test for detecting KPC and OXA-48 producers but has low sensitivity for MBL producers[21,22] and low specificity due to false positives from ESBL or AmpC β-lactamase producers with porin loss.[22-24] Also, it requires long incubation period of 24 hours. CNP test is a rapid chromogenic test which gives results in 2 hours with approximately 100% sensitivity and specificity for detecting carbapenemases in Enterobacteriaceae. But its cumbersome procedure requires bacterial extract. Modified strip CNP test is a modification of the CNP test and can be performed directly on bacterial culture instead of bacterial extract. The sensitivity and specificity of this test is reported to be equivalent to the original CNP test.[25,26]
The objective of present study was to evaluate the modified strip CNP test against the MHT for detection of carbapenemase production on CRE isolates.
Materials and Methods
Present study was performed in the Microbiology department of tertiary care teaching hospital of North India from September 2018 to August 2019 with due permission from Institutional Ethical Committee.
Various clinical specimens like pus, blood, sputum, throat swab, endotracheal tube aspirate, CSF, urine, body fluids, etc. obtained from various medical and surgical wards, ICU and OPD were included in the study. These samples were further processed for identification of organism as per routine laboratory protocols. Organisms identified as members of Enterobacteriaceae were screened for carbapenem resistance by Kirby Bauer disk diffusion method following CLSI 2018 guidelines using Meropenem 10 μg disk. Isolates which show inhibition zone diameter of ≤19 mm were considered as screening test positive and named as carbapenem-resistant Enterobacteriaceae (CRE).
One hundred seven CRE isolates were further evaluated for carbapenemase production by MHT and Modified strip CNP test.
MHT[27] is performed as per recommendations of CDC. MHT Positive test has a clover leaf-like indentation of the Escherichia coli 25922 growing along the test organism growth streak within the disk diffusion zone.
Modified strip CNP test[25] is a colorimetric test based on detection of hydrolysis of the β-lactam ring of a carbapenem (imipenem) by carbapenemase enzyme produce by bacteria. Hydrolysis of imipenem acidifies the medium, changing the color of the pH indicator (phenol red solution) from red to yellow. In present study, CNP test was modified by using paper strip as medium. In addition, pharmaceutical imipenem + cilastatin powder was used as a substrate in place of standard imipenem powder further reducing the cost. This test was performed directly on bacterial culture instead of bacterial extract making it easy as compared with original CNP test. Reagents used were same as original CNP test; Solution A (phenol red, ZnSO4.7H2O, pH 7.8) and Solution B (Solution A + 12 mg/mL of imipenem). A Whatman filter paper is cut into square strips of size 15 × 15 mm. One strip was labeled as control strip and other as test strip. Control strip was moistened with 50 μL of solution A while Test strip was moistened with 50 μL of solution B. Subsequently one calibrated loop (1 μL) of test organism grown on Mueller Hinton agar at 37°C for 18 to 24 hours was directly applied on the control and test strip by rubbing in circular fashion of approximately 5 to 7 mm in diameter. Results were read in 1 to 5 minutes. Isolates giving yellow color on the test strip with more intensity than on the control strip were considered as carbapenemase producers (►Fig. 1).

- Modified strip Carba NP test.
K. pneumoniae ATCC BAA1705 strain was used as positive control and K. pneumoniae ATCC BAA1706 as negative control.
Statistical Analysis
Statistical Package for the Social Sciences (SPSS) software, IBM India; version V26 was used for statistical analysis. The data collected was qualitative and expressed as proportions or percentage. To ensure the results obtained are statistically significant within a 95% level of confidence (with p-value 0.05), systematic hypothesis tests like Z-test and Chi-square test have been performed. Results were analyzed to determine if there was any significant difference in efficiency of MHT and Modified strip CNP test for early detection of CPE. Crosstabs were used to demonstrate the relationship between the two test types MHT and Modified strip CNP test. Cohen's kappa coefficient (κ) was used to measure inter-rater reliability (measure of agreement between two test types).
Results
A total of 107 CRE isolates were included in this study. Majority of CRE was isolated from age group 21 to 40 years (36%) followed by 41 to 60 years (23%) with male preponderance over female patients (►Fig. 2). Maximum number of CRE were isolated from samples received from wards 55 (52%) followed by intensive care unit (ICU) 27 (25%) and outdoor units 25 (23%). CRE was isolated in maximum number from urine samples 44 (41%) followed by ET secretion 27 (25%) (►Fig. 3).

- Distribution of CRE among various age groups. CRE, carbapenem-resistant Enterobacteriaceae.

- Distribution of CRE in various clinical samples included in study population. CRE, carbapenem-resistant Enterobacteriaceae.
Out of 107 CRE isolated, the most common species isolated were Enterobacter species 50 (47%) followed by E. coli 32 (30%), Klebsiella species 18 (17%), Citrobacter species 4 (4%), and Proteus species 3 (3%) (►Table 1).
| Isolates | No. of CRE isolates | MHT positive | CNP positive | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Escherichia coli | 32 | 11 | 34 | 25 | 78 |
| Enterobacter aerogenes | 30 | 12 | 40 | 27 | 90 |
| Enterobacter cloacae | 20 | 06 | 30 | 18 | 90 |
| Klebsiellaspp. | 18 | 13 | 72 | 18 | 100 |
| Citrobacterspp. | 4 | 03 | 75 | 4 | 100 |
| Proteus mirabilis | 2 | 01 | 50 | 1 | 50 |
| Proteus vulgaris | 1 | 00 | 0 | 1 | 100 |
| Total | 107 | 46 | 43 | 94 | 88 |
Abbreviations: CNP, Carba NP test; CRE, carbapenem-resistant Enterobacteriaceae; MHT, modified Hodge test.
Out of 107 CRE isolates, 46 (43%) isolates were phenotypically confirmed as carbapenemase producer by MHT and 94 (88%) were confirmed by Modified strip CNP test (at significance level of α = 0.5, Chi-square value with 1 degree of freedom is 47.592, Z-value is 6.89, p-value< 0.05 for both Chi-square test and Z-test, significantly) (►Table 1; ►Fig. 4), 38 (36%) isolates showed carbapenemase production by both MHT and CNP test, five (5%) isolates were both MHT and CNP test negative, eight (7%) were MHT positive but CNP test negative and 56 (52%) were MHT negative but CNP test positive (►Table 2). Measure of agreement (kappa value) is 0.081 which concludes no agreement between two test types: MHT and Modified strip CNP test.
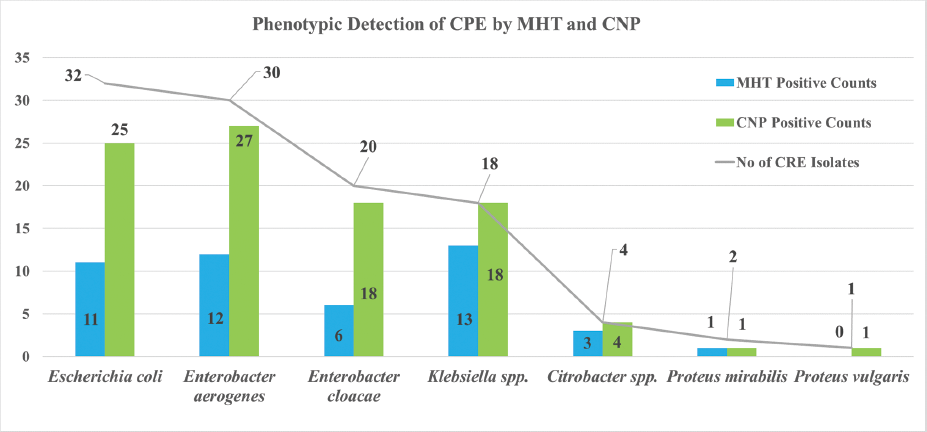
- Phenotypic detection of CPE by MHT and Modified strip CNP test. CNP, Carba NP; CPE, carbapenemase producing Enterobacteriaceae.
| isolates | CRE | MHT positive | CNP positive | MHT and CNP positive | MHT positive-CNP negative | MHT negative and CNP positive | MHT and CNP negative |
|---|---|---|---|---|---|---|---|
| Escherichia coli | 32 | 11 (34%) | 25 (78%) | 7 (22%) | 4 (13%) | 18 (56%) | 3 (9%) |
| Enterobacter aerogenes | 30 | 12 (40%) | 27 (90%) | 10 (33%) | 2 (7%) | 17 (57%) | 1 (3%) |
| Enterobacter cloacae | 20 | 6 (30%) | 18 (90%) | 5 (25%) | 1 (5%) | 13 (65%) | 1 (5%) |
| Klebsiellaspp. | 18 | 13 (72%) | 18 (100%) | 13 (72%) | 0 (0%) | 5 (28%) | 0 (0%) |
| Citrobacterspp. | 4 | 3 (75%) | 4 (100%) | 3 (75%) | 0 (0%) | 1 (25%) | 0 (0%) |
| Proteus mirabilis | 2 | 1 (50%) | 1 (50%) | 0 (0%) | 1 (50%) | 1 (50%) | 0 (0%) |
| Proteus vulgaris | 1 | 0 (0%) | 1 (100%) | 0 (0%) | 0 (0%) | 1 (100%) | 0 (0%) |
| Total | 107 | 46 (43%) | 94 (88%) | 38 (36%) | 8 (7%) | 56 (52%) | 5 (5%) |
Abbreviations: CNP, Carba NP test; CRE, carbapenem-resistant Enterobacteriaceae; MHT, modified Hodge test.
Discussion
Carbapenems are commonly used to treat infections caused by ESBL and/or AmpC β-lactamases producing and other MDR Enterobacteriaceae. Although they are stable to hydrolysis by most β-lactamases, their usage as the last resort antibiotics was seriously compromised by the appearance of new class of bacterial enzymes capable of inactivating carbapenems, called as carbapenemases.[28]
Resistance to carbapenem antimicrobials is either due to expression of carbapenemases or due to combined mechanisms of resistance (overexpression of broad-spectrum β-lactamases together with efflux pumps, impermeability); however, carbapenemases production represents the most important mechanism of resistance in Enterobacteriaceae, associated with multi- or pan-drug resistance.
Currently, KPC, MBL, and OXA-48 carbapenemases are considered as major threat in Enterobacteriaceae. They are independently disseminated between different nations worldwide. In India, very few reports on KPC-producing isolates exist. India being the main identified reservoir for NDM type MBL carbapenemases.[12-14]
The last decade is marked with the rapid spread of carbapenemases-producing Enterobacteriaceae worldwide. These infections are often associated with MDR and account for high mortality in patients. Therefore, early detection of CPE is highly crucial not only to guide proper treatment but also to prevent spread of Carbapenem resistance in the society.
In present study, resistance to carbapenem was seen in all age groups and in both the genders. Though in less number, pediatric and geriatric populations were also found to be affected. CRE was isolated from both nosocomial as well community-acquired infection; however, nosocomial spread seems to be the main mode of dissemination (77% CRE was isolated from IPD and 23% from OPD). Similar findings were seen in study by Nair and Vaz[29] in which most of the CRE isolates were detected in patient samples from the wards (42%) and the ICU (26%) followed by OPD patients (19%). Chauhan et al[30] and Pandurangan et al[31] also have same observation. Thus, to prevent CRE spread in hospital settings, measures like hand hygiene, contact precautions, minimizing the use of invasive devices, patient and staff cohorting should be given due importance. CRE isolation from samples received in outdoor patient is also of major concern as Enterobacteriaceae mainly colonizes human gut flora; CRE strains can spread rapidly into the community via the fecal oral route. Therefore, screening the contacts of CRE patients and active surveillance testing could possibly help to curtail down the CRE transmission in the community.
CRE was isolated in maximum number from urine (41%) samples followed by ET secretion (25%), pus (11%), sputum (9%), CSF (6%), and blood (3%). The results are consistent with studies done by many authors.[29-32] In all these studies CRE was isolated mainly from urine samples followed by respiratory samples.
In present study, dissemination of carbapenem resistance was seen among all the species of Enterobacteriaceae family as out of 107 CRE isolated, 50 (47%) were Enterobacter species, 32 (30%) were E. coli, 18 (17%) were Klebsiella species, four (4%) were Citrobacter species, and three (3%) were Proteus species. Similarly, studies done by other authors also reported interspecies dissemination of carbapenem resistance like Datta et al[33] reported carbapenem resistance in 62.12% of K. pneumoniae, 7.58% of Klebsiella oxytoca, and 16.6% of E. coli. Bartolini et al[34] reported maximum carbapenem resistance in Klebsiella species (88%) followed by Enterobacter species (8%) and E. coli (4%). Pandurangan et al[31] observed CRE in E. coli (61%), K. pneumoniae (21.5%), and Enterobacter cloacae (6%). This may be because genes carrying CRE are located on plasmids and transposons causing rapid dissemination of carbapenem resistance. These studies also suggested that CRE were mainly seen in Enterobacter species, E. coli, and Klebsiella species. CDC (2019) has also incorporated these three species in their case definition for surveillance of CP-CRE cases.[35]
CRE isolates were subjected to Modified strip CNP test and MHT for detection of carbapenemase production. Out of 107 CRE isolates, 46 (43%) were confirmed phenotypically as carbapenemase producer by MHT and 94 (88%) were confirmed by Modified strip CNP test. There is statistically significant difference in efficiency of MHT and Modified strip CNP test for detection of carbapenemase producing CRE isolates (p-value <0.05). Modified strip CNP test was more sensitive than MHT for detecting CP-CRE. Our results are consistent with the studies conducted by many authors. The study by Datta et al[33] observed CNP test (86.4% [114/132] positive) shows better result as compared with MHT (75% [99/132] positive). Similarly, in studies done by Vasoo et al[36] and Yamada et al[37] CNP test was found to be more sensitive and specific for detecting CPE as compared with MHT. This may be because Indian subcontinent is endemic for MBL type of carbapenemases mainly NDM.[10,12-14] and several studies reported MHT has low sensitivity for detecting MBL producers. Bartolini et al[34] observed MHT is 100% sensitive for the detection of KPC but has low sensitivity for VIM and was not able to detect NDM-1. Similarly, Girlich et al[21] reported that MHT has an excellent sensitivity for detecting KPC and OXA-48 carbapenemases but has low sensitivity (50%) for NDM-1 producers. EUCAST guidelines[38] mentioned MHT has low sensitivity for MBL detection. This may be likely due to zinc supplementation in reagents used for Modified strip CNP test which is lacking in MHT.[21]
In present study, measure of agreement (kappa) was 0.081(no agreement between the two tests). It was in accordance with Datta et al[33] who also reported measure of agreement (kappa) value less than 0.001. Out of 107 CRE isolates, 38 (36%) isolates showed carbapenemase production by both MHT and CNP tests. Fifty-six isolates (52%) were CNP test positive but MHT negative, five (5%) isolates were both MHT and CNP test negative that is they were not identified as carbapenemase producer by either of the test. It may be because these isolates were resistant to carbapenems because of mechanism other than carbapenemase production. Eight (7%) isolates were MHT positive but CNP test negative; this may be because of two reasons, either MHT gives false positive results as MHT suffers from poor specificity due to false positive results for isolates producing ESBL or AmpC β-lactamase (AmpC) combined with porin loss as reported by many authors[22-24] or modified strip CNP test has given false negative results as CNP test has low sensitivity for OXA-48 producers.[39]
CRE isolates are not only resistant to beta lactam antibiotics but also show significant cross resistance between different classes of antibiotics as plasmids carrying genes for carbapenem resistance also carry MDR determinant genes. Infections caused by CRE are resistant to most of the available antibiotics; they are difficult to treat and are therefore associated with major therapeutic failures, increased duration of hospital stays with increased cost of treatment, and high mortality rate. Also, resistance to carbapenems due to carbapenemase production is rapidly transmissible between different species of Enterobacteriaceae, thus early detection of CPE holds high merits to prevent spread of CRE.
Modified strip CNP test shows promising results as compared with MHT. It is simple, inexpensive, reproducible test which is easy to perform and interpret and give rapid results in less than 5 minutes. It has high degree of sensitivity and specificity comparable to molecular test. On the other hand, MHT is a cumbersome procedure; results are difficult to interpret and require 24-hour incubation time. Its sensitivity is low especially for MBL producers and also has low specificity.
Conclusion
CP-CRE have high tendency to spread in the community and are associated with high morbidity and mortality. Modified strip CNP test is a practical solution for early detection of CP-CRE particularly in low resource countries like India which are large reservoir of carbapenemase producers. It can contribute to formulate better treatment plan to avoid therapeutic failures and to design logical infection control policies to prevent further dissemination of carbapenemase producers in the community thus shifting the paradigm of carbapenem resistance.
Authors’ Contribution List
N.P. undertook the research work under the guidance of co-authors, reviewed the literature, did data collection, data analysis, and interpretation, and prepared manuscript. N.V. conceptualized and designed the research work, approved the protocol, guided the research process, provided intellectual input, and proofread the manuscript. S.S. reviewed the literature and did data analysis and interpretation. B.S. provided intellectual input and guidance throughout the research process, did manuscript correction, and revised manuscript for important intellectual content
Financial Disclosure
None.
Conflict of Interest
None.
References
- Koneman's color atlas and textbook of Diagnostic Microbiology, Characteristics for Presumptive Identification of Enterobacteriaceae. (7th). Philadelphia, PA: Wolters Kluwer; 2017. p. :685-689.
- [Google Scholar]
- Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(01):42-51.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J Clin Microbiol. 2011;49(08):2924-2932.
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55(11):4943-4960.
- [CrossRef] [PubMed] [Google Scholar]
- Novel carbapenem-hydrolyzingβ-lactamase, KPC1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(04):1151-1161.
- [CrossRef] [PubMed] [Google Scholar]
- Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. ISBN 2017 978–92–4-155017–8. Available at: https://www.who.int/infection-prevention/publications/guidelines-cre/en/. Accessed November 17, 2019
- [Google Scholar]
- Bailey & Scotts Diagnostic Microbiology, Expanded Spectrum Cephalosporin Resistance & Carbapenemase Resistance. (13th). St. Louis: Elsevier; 2014. p. :326.
- [Google Scholar]
- Metallo-β-lactamases: a last frontier forβ-lactams? Lancet Infect Dis. 2011;11(05):381-393.
- [CrossRef] [PubMed] [Google Scholar]
- The emerging NDM carbapenemases. Trends Microbiol. 2011;19(12):588-595.
- [CrossRef] [PubMed] [Google Scholar]
- OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67(07):1597-1606.
- [CrossRef] [PubMed] [Google Scholar]
- The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8(04):460-469.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J Clin Microbiol Infect Dis. 2015;34(03):467-472.
- [CrossRef] [PubMed] [Google Scholar]
- blaKPC gene detection in clinical isolates of carbapenem resistant Enterobacteriaceae in a tertiary care hospital. J Clin Diagn Res. 2013;7(12):2736-2738.
- [CrossRef] [PubMed] [Google Scholar]
- Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(09):1503-1507.
- [CrossRef] [PubMed] [Google Scholar]
- Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(01):119-123.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of a DNA microarray for the rapid detection of extended-spectrumβ-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA48, VIM, IMP and NDM) J Antimicrob Chemother. 2012;67(08):1865-1869.
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49(09):3222-3227.
- [Google Scholar]
- Comparison of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM2, NDM1, SIM1, KPC1, OXA23, and OXA-51 carbapenemase-producing Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn Microbiol Infect Dis. 2013;77(03):227-230.
- [CrossRef] [PubMed] [Google Scholar]
- Rapidec Carba NP Test for rapid detection of carbapenemase producers. J Clin Microbiol. 2015;53(09):3003-3008.
- [CrossRef] [PubMed] [Google Scholar]
- Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50(02):477-479.
- [CrossRef] [PubMed] [Google Scholar]
- Cloverleaf test (modified Hodge test) for detecting carbapenemase production inKlebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65(02):249-251.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007;45(08):2723-2725.
- [CrossRef] [PubMed] [Google Scholar]
- New Delhi metallo-β-lactamase (NDM-1)-producing Klebsiella pneumoniae: case report and laboratory detection strategies. J Clin Microbiol. 2011;49(04):1667-1670.
- [CrossRef] [PubMed] [Google Scholar]
- Modification and evaluation of the Carba NP test by use of paper strip for simple and rapid detection of carbapenemase-producing Enterobacteriaceae. World J Microbiol Biotechnol. 2016;32(07):117.
- [CrossRef] [PubMed] [Google Scholar]
- Rapid detection of carbapenemase production in Enterobacteriaceae by use of a modified paper strip Carba NP method. J Clin Microbiol. 2017;56(01):e01110-e01117.
- [CrossRef] [PubMed] [Google Scholar]
- Modified Hodge test for carbapenemase detection in Enterobacteriaceae. Available at: https://www.cdc.gov/HAI/pdfs/labSettings/HodgeTest_Carbapenemase_Enterobacteriaceae.pdf. Accessed November 17, 2019
- [Google Scholar]
- Prevalence of carbapenem resistant Enterobacteriaceae from a tertiary care hospital in Mumbai, India. J Microbiol Inf Dis. 2013;3(04):207-210.
- [CrossRef] [Google Scholar]
- Evaluation of phenotypic tests for detection of Klebsiella pneumoniae carbapenemase and metallo-beta-lactamase in clinical isolates of Escherichia coli and Klebsiella species. Indian J Pathol Microbiol. 2015;58(01):31-35.
- [CrossRef] [PubMed] [Google Scholar]
- Phenotypic detection methods of carbapenemase production in Enterobacteriaceae. Int J Curr Microbiol Appl Sci. 2015;4(06):547-552.
- [Google Scholar]
- In-house standardization of Carba NP test for carbapenemase detection in gram negative bacteria. Int J Curr Microbiol Appl Sci. 2018;7(01):2876-2881.
- [CrossRef] [Google Scholar]
- A comparative study of modified Hodge test and Carba NP test for detecting carbapenemase production in gram-negative bacteria. Med J Dr D.Y. Patil Uni. 2017;10:365-369.
- [CrossRef] [Google Scholar]
- Comparison of phenotypic methods for the detection of carbapenem non-susceptible Enterobacteriaceae. Gut Pathog. 2014;6:13.
- [CrossRef] [PubMed] [Google Scholar]
- Case definition for surveillance of CP-CRE. 2018 Available at: https://wwwn.cdc.gov/nndss/conditions/carbapenemase-producing-carbapenem-resistant-enterobacteriaceae/case-definition/2018/. Accessed November 17, 2019
- [Google Scholar]
- Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol. 2013;51(09):3097-3101.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of the Modified-Hodge test, Carba NP test, and carbapenem inactivation method as screening methods for carbapenemase-producingEnterobacteriaceae. J Microbiol Methods. 2016;128:48-51.
- [CrossRef] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. . 2017;2:1-43.
- [Google Scholar]
- Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57(09):4578-4580.
- [CrossRef] [PubMed] [Google Scholar]





