Translate this page into:
Profile of Nontuberculous Mycobacteria in Patients Suspected of Tuberculosis and Drug-Resistant Tuberculosis
Address for correspondence Bharti Malhotra, MBBS, MD, Department of Microbiology, SMS Medical College, Jaipur, Rajasthan, 302004 India (e-mail: drbhartimalhotra@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
Infections due to nontuberculous mycobacteria (NTM) is increasing globally and may present as drug-resistant tuberculosis (DRTB). In India, data on NTM prevalence and species diversity is limited. Present study was conducted to detect the prevalence and profile of NTM among patients suspected of DRTB using paraffin slide culture (PSC)and mycobacteria growth indicator tube (MGIT) culture methods for isolation of NTM.
Material and Method
A total of 2,938 samples suspected of TB/DRTB were cultured on PSC and MGIT960. Species identification of mycobacterial isolate was done by sequencing of 16s ribosomal RNA gene.
Result
Among 2938 samples, 35 (1.19%) were found positive for NTM by PSC and 9 (0.30%) were found positive by MGIT. The diversity of NTM species was high (13 species). Out of 35 NTM isolates by PSC, maximum 34.29% (12) isolates were found to be Mycobacterium fortuitum, followed by 11.43% (4) Mycobacterium abscessus and Mycobacterium chelonae, and 42.85% (15) were other species viz. 8.57% (3) were Mycobacterium intracellulare and Mycobacterium kansasii, 5.71% (2) were Mycobacterium peregrinum, and 2.85% (1) were Mycobacterium flavescens, Mycobacterium farcinogenes, Mycobacterium moriokanese, Mycobacterium wolinskyi, Mycobacterium simiae, Mycobacterium goodii, and Mycobacterium terrae each. Coinfection of Mycobacterium tuberculosis(MTB) and NTM was found in 60% (21) samples.
Conclusion
Prevalence of NTM was low among multidrug resistant tuberculosis/TB suspected patients, similar to other studies done in India. PSC was found better than MGIT for the isolation of NTM, though poor separation of NTM and MTB on subculture may have led to false negativity in cases of coinfection. About 13 species were isolated; M. fortuitum was the most common of all. Since coinfection of NTM and TB can also occur, samples of patients suspected of NTM should be cultured on PSC even if positive for MTB.
Keywords
nontuberculous mycobacteria (NTM)
paraffin slide culture (PSC)
mycobacterium tuberculosis (MTB)
Introduction
Nontuberculous mycobacteria (NTM) are opportunistic pathogens that cause surgical-site infections, postinjection abscesses, osteomyelitis, catheter-related bloodstream infections, and central nervous system infections, mostly affecting patients with the pre-existing pulmonary disease such as chronic obstructive pulmonary disease (COPD) or tuberculosis (TB), or immunocompromised patients with HIV infection, leukemia, or those patients on immunosuppressive drugs.1,2 The exact burden of NTM disease is not known because it is not a reportable disease worldwide including India, but now NTM infections are showing increasing trend in India.3 The clinic radiological signs and symptoms of both NTM and Mycobacterium tuberculosis (MTB) are quite similar; thus, NTM is easily misdiagnosed as MTB, or as multidrug-resistant tuberculosis (MDRTB) and extensively drug-resistant tuberculosis (XDRTB) that pose challenge for effective treatment.3 NTM isolation rates range from 0.5 to 8.6% in India.4 A recent study from India reported increase in prevalence of NTM from 1.0% in 2005 to 3.5% in 2008; 88.6% of these NTM were clinically relevant.5 The isolation rates of NTM vary widely depending on population and geographic location.6 Infections with pathogenic NTM such as Mycobacterium marinum, Mycobacterium fortuitum, and Mycobacterium chelonae have been reported from India.7
Paraffin baiting has been used for isolating NTM and Nocardia8,9 as NTM utilizes paraffin wax as the sole source of carbon, while MTB is not able to utilize paraffin wax as the sole source of carbon and does not grow in the paraffin slide culture (PSC). This property helps in isolating NTM from clinical specimens as this property of paraffin metabolism is not found among other human pathogens.9
It is important to isolate and identify NTM because of its pathogenic potential and higher resistance to antituberculosis drugs.10 Phenotypic identification of NTM species is based on a panel of biochemical tests,11which have many drawbacks such as slow turnaround time, difficult to interpret, low specificity, and limited accuracy; laboratories are now using alternative rapid methods for species identification such as DNA probes (AccuProbe, Gen-Probe, San Diego, California, United States),polymerase chain reaction (PCR)-restriction enzyme analysis of the hsp65 gene,12 or sequencing of the 16S ribosomal DNA (rDNA).13 Use of 16S ribosomal RNA (rRNA) gene has now become a “gold standard” for the identification of NTM.14
Hence, present study was conducted to investigate the prevalence and profile of NTM among DRTB suspected patients using mycobacteria growth indicator tube (MGIT) and PSC methods for isolation and 16S rRNA gene sequencing for species identification.
Procedure
Study Setting
Study was performed in Mycobacteriology Laboratory, Department of Microbiology, SMS Medical College, Jaipur, Rajasthan from August 2016 to August 2017. Samples of presumptive DRTB and TB suspected patients were included in the study. This study was approved by the Institutional Ethical Committee.
Inclusion Criteria
Pulmonary samples with more than 5 mL volume and extrapulmonary samples with more than 2 mL volume were included in the study.
All samples received in laboratory having adequate volume were included in the study. Among 2,938 samples enrolled in the study, 1,612 were from DRTB suspect and1,326 were from pulmonary and extrapulmonary TB suspects. All clinical samples, pulmonary (sputum, bronchoalveolar lavage) and extrapulmonary (gastric aspirate, pleural fluid, pus and endometrial biopsy), from males and females of all age groups were included in this study. Handling of MGIT cultures and PSC cultures were done in biosafety level 3 (BSL3) laboratory, while rest all processes were done in BSL2 laboratory as recommended for MTB.
Statistical Analysis
All data were entered in Microsoft Excel version 10 and analyzed by SPSS version 21. Chi-squared test was applied to find statistical significance.
Sample Collection
Samples were collected in sterile, screw cap containers from various districts of Rajasthan, stored in refrigerator at the districts for 1 to 5 days and transported to the laboratory by courier or hand delivered maintaining cold chain. On receipt of samples at laboratory, they were processed immediately or after refrigeration at 4°C overnight.15 For PSC, the deposits of processed samples were stored in–20°C and inoculated once in 7 to 10 days; rest all work was done daily as per Revised National Tuberculosis Control Program (RNTCP) guidelines.
Sample Processing and Culture
All the samples were subjected to smear microscopy by Ziehl–Neelsen staining method, decontamination byN-acetyl-L-cysteine-sodium hydroxide method (with final NaOH concentration of 4%) as per RNTCP guidelines.10 The concentrated sediment (1–1.5mL) was resuspended in 1 to 2 mL of phosphate buffer and processed for MGIT culture (0.5mL) and line probe assay (LPA) (0.1mL) as per RNTCP guidelines.16,17 Rest of deposit was stored at–20°C; 0.5 mL of the stored deposit of all samples was inoculated in PSC.9 Smear-positive DRTB suspected samples were processed directly for LPA from sample, MTB growth from MGIT was processed for LPA if direct LPA/GeneXpert results were not available.17,18 Samples (1mL) of TB suspects were processed directly for GeneXpert as per protocol.18 All the processing done in this study is shown in ►Fig. 1.

- Workflow chart (STROBE Guideline). AFB, acid-fast bacilli; DRTB, drug-resistant tuberculosis; LJ, Lowenstein-Jensen medium; LPA, line probe assay; MGIT, mycobacteria growth indicator tube; MTB, Mycobacterium tuberculosis; NTM, nontuberculous mycobacteria; PSC, paraffin slide culture; STROBE, Strengthening The Reporting of Observational studies in Epidemiology
PSC
Briefly, 0.5 mL of processed sample was inoculated into 4.5 mL sterile Czapek broth medium (Czapek broth constituents: NaNO3—3.0 g, K2HPO4—1.0, MgSO4H2O—0.5 g, KCl—0.5 g, FeSO4—0.01 g) with added BACTEC PANTA Plus (Becton Dickinson) (polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin) to reduce contamination, into which dipped sterile paraffin coated glass slides were dipped, incubated at 37°C, and checked daily for growth. The sample was considered as negative when no growth was seen on paraffin-coated slide till 6 weeks. When growth appeared on slide, it was taken out from the broth, colonies were scratched and subcultured on Lowenstein-Jensen medium (LJ) and this paraffin coated slide was stained by Kinyon’s staining method by placing it into a tube containing Kinyoun carbol fuchsin.9
MGIT Culture
Deposit (0.5 mL) of all processed samples were inoculated in MGIT tubes (Becton Dickinson) as per protocol.16 MGIT culture-positive isolates were processed for acid-fast bacilli (AFB) smear preparation, AFB-positive isolates were subjected to MPT64 antigen test as mentioned below, MPT64 antigen-positive growth was subcultured on LJ, and samples positive for MTB were subjected to LPA if direct LPA and GeneXpert results (positive) were not available.
SD BIO-LINE TB Ag MPT64 Rapid
AFB-positive culture isolates from MGIT and LJ were subjected to SD BIO-LINE TB Ag MPT64 rapid test (Standard Diagnostics, Korea). MPT64 Ag discriminates between MTB and NTM (NTM is negative for MPT 64 antigen).19
LPA
Smear-positive DRTB suspected samples were subjected directly for LPA Hain GenoType MTBDRplus assay (Hain Lifescience GmbH, Nehren, Germany) as per protocol.17
GeneXpert
Pulmonary and extrapulmonary TB suspected samples were subjected to GeneXpert (Cepheid) for MTB and rifampicin resistance detection as per protocol.18
DNA Extraction
DNA was extracted from AFB smear-positive sputum deposit/culture isolate obtained in MGIT/LJ using GenoLyse Version 1.0 (HAIN Lifescience GmbH, Nehren, Germany) according to manufacturer's instructions20 for carrying out direct/indirect LPA and sequencing. DNA was stored for future use at –20°C.
16s rRNAgene sequencing
DNA extracted from NTM isolates was subjected to PCR.14 The reaction mix consisted of 25µL Hot Start Taq Mastermix (QIAGEN), 2 µL forward primer, 2 µL reverse primer, 18 µL nuclease-free water, and 3µL sample DNA. Cycling conditions were as follows:1 cycle of 94°C for 15 minutes, 35 cycles each of denaturation, annealing, and extensionat at 94°C for 30 seconds,56°C for 30 seconds, and 72°C for 45 seconds each, respectively, followed by 1 cycle of final extension at 72°C for 7 minutes. The PCR products were analyzed by gel electrophoresis. Sequencing was done by using forward primer. Species identification was done by 16s rRNA gene sequencing on the ABI 3500 Genetic Analyzer using the BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystem) as per the protocol.The sequence was aligned and compared with those available in the GeneBank database by BLAST (Basic Local Alignment Search Tool) analysis.21 Nucleotide sequences obtained from the study are available in the GenBank database (accession numbers MH882687- MH882721).
Results
Among 2,938 samples cultured on PSC and MGIT 960, growth was observed in 70 (2.3%) samples by PSC, 35 (1.19%) were NTM species, 8 (0.27%) were Nocardia species, and in 27 (0.9%) samples contamination was found, MTB was not grown on PSC. On MGIT, 1,247 (42.44%) samples were reported as MTB and 9 (0.30%) as NTM. The nine NTM species isolated by MGIT were same as that isolated by PSC. In PSC, 26 additional NTM were isolated than MGIT culture (►Table 1, ►Fig. 2).
| Patient criteria | Total | MGIT culture positive for MTB | MGIT culture negative for MTB | NTM isolated by MGIT | MGIT contamination | NTM isolated by PSC |
|---|---|---|---|---|---|---|
| Smear positive DRTB suspected patients | 840 | 800 (95.23%) | 20 (2.38%) | 6a (0.71%) | 19 (2.26%) | 17 (2.02%) |
| Smear-negative DRTB suspected patients | 772 | 310 (40.15%) | 400 (51.81%) | 2b (0.25%) | 61 (7.90%) | 11 (1.42%) |
| Smear-positive TB suspected patients | 102 | 94 (92.15%) | 6 (5.88%) | – | 2 (1.96) | 2 (1.96%) |
| Smear-negative TB-suspected patients | 1224 | 43 (3.51%) | 1,151 (94.03%) | 1 (0.08%) | 29 (2.36) | 5 (0.40%) |
| Total | 2,938 | 1,247 (42.44%) | 1,577 (53.67%) | 9 (0.30%) | 111 (3.77%) | 35 (1.19%) |
Abbreviations: DRTB, drug-resistant tuberculosis; MGIT, mycobacteria growth indicator tube; MTB, Mycobacterium tuberculosis; NTM, nontuberculous
mycobacteria; PSC, paraffin slide culture.
Note: Twenty-six additional NTM were isolated by PSC than MGIT culture.
aFive patients had NTM and MTB coinfection.
bOne patient had NTM and MTB coinfection.
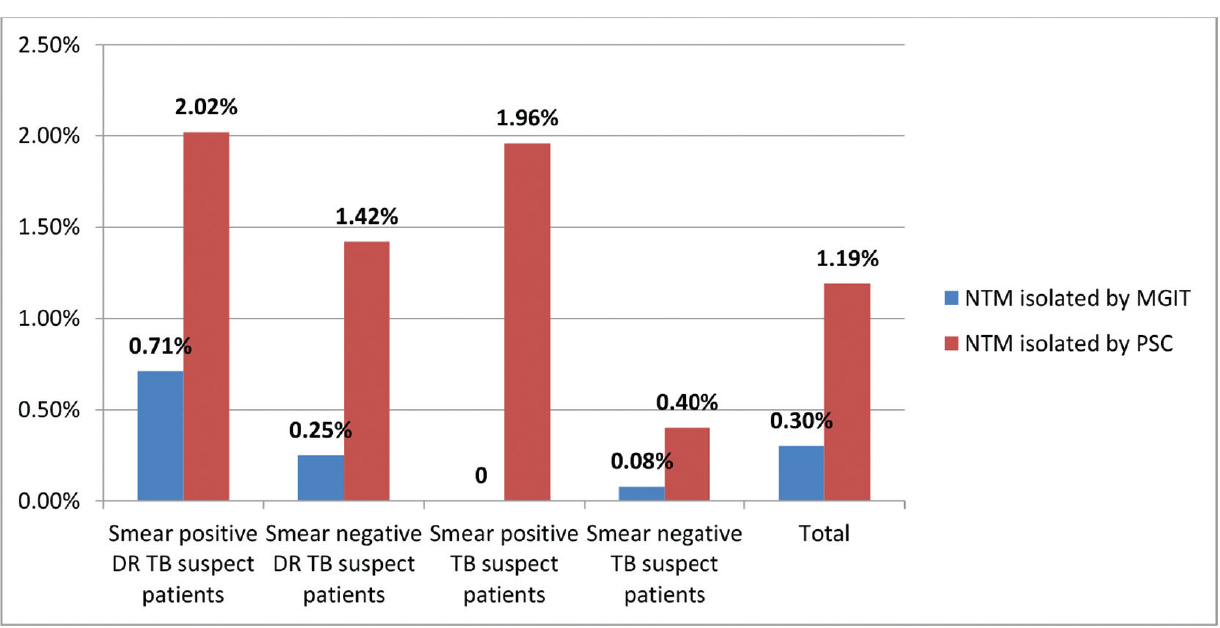
- Nontuberculous mycobacteria (NTM) isolation by mycobacteria growth indicator tube (MGIT) and paraffin slide culture (PSC) in different group of patients.
Among nine NTM isolated by MGIT, three NTM (1 was invalid by direct LPA) were identified directly as NTM from MGIT culture, while six NTM were identified after subculture on LJ media from MGIT that was positive for MTB.
Out of 1,612 DRTB suspected samples, 1,149 (71.27%) samples were positive for MTB and among 1,326 TB suspected samples, 135 (10.18%) samples were MTB positive by GeneXpert and 137 (10.33%) by MGIT. Among 1149 DRTB suspects, 10% samples were MDR, 10.69% were isoniazid resistant, 2.69% were rifampicin resistant, and 76.52% were rifampicin and isoniazid sensitive by LPA. Among 1,326 TB suspects, 6.66% samples were rifampicin resistant by GeneXpert. Among 1,286 total MTB-positive samples, 155 (12.05%) samples were MDR.
Twenty-one (60%) patients had coinfection with MTB (identified by growth in MGIT or positive by GeneXpert) and NTM (isolated on PSC/MGIT) and remaining 14 (40%) had only NTM infection (►Fig. 3). Among the coinfection cases, 90.4% (19) were in pulmonary samples and 9.5% (2) in extrapulmonary samples. Among 19 pulmonary samples, 31.5% (6) samples were rifampicin resistant, 5.2% (1) were isoniazid resistant, and 42.1% (8) were MDR. Among two extrapulmonary samples, 50% (1) samples were rifampicin resistant and 50% (1) were rifampicin sensitive.

- Coinfection of Mycobacterium tuberculosis along with nontuberculous mycobacteria(NTM). DRTB, drug-resistant tuberculosis; MGIT, mycobacteria growth indicator tube; MTB, Mycobacterium tuberculosis.
Positivity for NTM was higher in males (1.27%) than females (1.02%). The predominant age group was > 55 years (25.7%) followed by 46 to 55 years (22.86%) age group, respectively.
A wide range of NTM species were isolated by PSC in our study. Out of 35 NTM isolates, maximum isolates were Mycobacterium fortuitum, (34.29%) followed by Mycobacterium abscessus (11.43%) and Mycobacterium chelonae (11.43%) and rest (42.85%) belonged to other species. M. fortuitum was the most common species, isolated from both DRTB suspected patients and TB suspected patients by PSC. However, among three NTM that were isolated by MGIT, three (33.3%) were found to be M. fortuitum and M. chelonae, each, two (22.2%) were M. abscessus, and one (11.1%) was Mycobacterium intracellulare (►Table 2).
| NTM species | DRTB suspects | TB suspectedsamples | Total | |||
|---|---|---|---|---|---|---|
| NTM species by PSC | NTM species by MGIT | NTM species by PSC | NTM species by MGIT | NTM by PSC | NTM by MGIT | |
| Mycobacterium fortuitum | 9 (32.14%) | 2 (25%) | 3 (42.87%) | 1 | 12 (34.29%) | 3 (33.3%) |
| Mycobacterium. abscessus | 4 (14.28%) | 2 (25%) | 0 | 0 | 4 (11.43%) | 2 (22.2%) |
| Mycobacterium chelonae | 4 (14.28%) | 3 (37.5%) | 0 | 0 | 4 (11.43%) | 3 (33.3%) |
| Mycobacterium intracellulare | 3 (10.71%) | 1 (12.5%) | 0 | 0 | 3 (8.57%) | 1 (11.1%) |
| Mycobacterium kansasii | 3 (10.71%) | 0 | 0 | 0 | 3 (8.57%) | 0 |
| Mycobacterium flavescens | 1 (3.57%) | 0 | 0 | 0 | 1 (2.85%) | 0 |
| Mycobacterium farcinogenes | 1 (3.57%) | 0 | 0 | 0 | 1 (2.85%) | 0 |
| Mycobacterium moriokanese | 1 (3.57%) | 0 | 0 | 0 | 1 (2.85%) | 0 |
| Mycobacterium wolinskyi | 1 (3.57%) | 0 | 0 | 0 | 1 (2.85%) | 0 |
| Mycobacterium simiae | 1 (3.57%) | 0 | 0 | 0 | 1 (2.85%) | 0 |
| Mycobacterium peregrinum | 0 | 0 | 2 (28.57%) | 0 | 2 (5.71%) | 0 |
| Mycobacterium goodii | 0 | 0 | 1 (14.28%) | 0 | 1 (2.85%) | 0 |
| Mycobacterium terrae | 0 | 0 | 1 (14.28%) | 0 | 1 (2.85%) | 0 |
| Total | 28 (100%) | 8 (100%) | 7 (100%) | 1 (100%) | 35 (100%) | 9 (100%) |
Abbreviations: DRTB, drug-resistant tuberculosis; MGIT, mycobacteria growth indicator tube; MTB, Mycobacterium tuberculosis; NTM, nontuberculous mycobacteria; PSC, paraffin slide culture.
The proportion of NTM in DRTB suspected patients and TB suspected patients was 1.73 and 0.52%, respectively, by PSC. This difference was found statistically significant (p-value = 0.003). By MGIT, only eight (0.27%) NTM were isolated from DRTB suspected patients and one (0.03%) from TB suspected patients.
In the present study, 82.8% (29/35) NTM were found in pulmonary samples and 17.1% (6/35) NTM were found in extrapulmonary samples (66.6% [4] from gastric aspirate and 33.3% [2] from pus).
Among 29 pulmonary samples, 10 were M. fortuitum, 4 were M. chelonae and M. abscessus, 3 were M. intracellulare and Mycobacterium kansasii, and 1 was Mycobacterium simiae, Mycobacterium farcinogenes, Mycobacterium moriokanese, Mycobacterium wolinsky, and Mycobacterium flavescens each. From six extrapulmonary samples, two species were M. fortuitum and Mycobacterium peregrinum and one species belongs to Mycobacterium goodii and Mycobacterium terrae.
Of 35 patients, 14.2% (5) had bronchiectasis, 17.1% (6) had bronchial asthma, 11.4% (4) had diabetes mellitus, 20% (7) had COPD, and 2.8% (1) had HIV. All these belonged to pulmonary samples. No predisposing condition was detected in 34.2% (12/35) patients.
On follow-up of patients/attendants telephonically, we observed that among the 35 patients with NTM, 7 (20%) patients (5 from DRTB suspects and 2 from TB suspect) expired during the treatment. Among the five expired DRTB suspected patients, three patients had coinfection with MTB and NTM. Ten (28.5%) (8 from DRTB suspects and 2 from TB suspects) patients have been cured after treatment among which seven patients had been coinfected with MTB and NTM. Four (11.4%) patients on treatment failed to recover, among which 2 (5.7%) had coinfection. There was no information available about 14 (40%) patients (►Table 3).
| No of patients | Clinical outcome | Coinfection present |
|---|---|---|
| 7 (20%) patients (5 DRTB/2 TB suspects) | 5 (14.2%) patients expired during treatment | In 3 (8.5%) patients (From DRTB suspects) |
| 10 (28.5%) patients (8 DRTB/2 TB suspects) | Cured after treatment | In 7 (20%) patients (5 from DRTB and 2 from TB suspects) |
| 4 (11.4%) patients (DRTB suspects) | On treatment fail to recover | In 2 (5.7%) patients |
| 14 (40%) patients | No information available | – |
Abbreviation: DRTB, drug-resistant tuberculosis.
Discussion
NTM have been considered to be contaminants or colonizers previously but since few years they have been identified as emerging pathogen, causing pulmonary and other infections among both immune-competent and immune-suppressed individuals6.The clinical profile of infections by NTM is quite similar to TB as a result they are often missed; moreover, specialized media is required for their growth that is not used in routine mycobacteriology laboratory and that increases the chance of missing NTM again. Lack of awareness among clinicians coupled with lack of laboratory capacity to diagnose these infections adds to problem of poor reporting of NTM.4
As per ATS guidelines published in 2007,1 NTM isolation from at least two separate pulmonary samples and isolation of NTM from sterile site or tissue is considered as confirmed NTM disease. Isolation of NTM once from nonsterile sources with compatible radiological and/or clinical findings is considered as highly probable NTM disease. Patients who do not fulfill the criteria for definite or highly probable NTM lung disease with the single isolation of NTM from clinical specimens are presumed to be suffering from possible NTM disease or colonization.
In the present study, we used the PSC method that is known to enhance the isolation of NTM.9
In our study 1.19% of NTM were isolated from DRTB and TB suspected samples by PSC. Wide variations in the prevalence of NTM were seen in studies from different parts of India ranging from 0.38% from Delhi,22 0.8% from Maharashtra,23 3.9% from south India,24 and 9.9% NTM in another study in Delhi.3 In some studies, NTM prevalence was low. A possible explanation for the low NTM disease prevalence in these studies can be the sample collection from a larger population as these healthcare facilities cover large number of districts in their respective states.
Yield of NTM was higher in PSC (1.19%) than MGIT (0.30%) in our study. In the previous studies, Narang et al9 and Ollar et al25 have reported PSC to be an excellent media for NTM and that no growth of MTB is obtained on PSC, similar results were obtained by us; however, no comparison of PSC and MGIT was available. As RNTCP is routinely using MGIT for culture of mycobacteria, it should be borne in mind that MGIT may not be able to isolate all the NTM. A study from India isolated 0.50% NTM from MGIT-positive cultures.26 There may be many possible reasons of lower isolation rates from MGIT, one is higher contamination in MGIT (4%) versus PSC (0.9%), presence of MTB-NTM coinfection due to which the colonies of NTM may not be identifiable and lay submerged under MTB growth, better sensitivity of PSC for NTM as only NTM are grown on it and MTB does not grow as a result even few colonies of NTM on PSC will be visible while on other hand subculture on LJ from positive MGIT may miss it. However, growth of only few colonies of NTM may not be clinically significant.
In our study, we observed that 5.8% samples of smear-positive TB suspects were negative by culture; similarly, culture positivity in smear negative samples was also low; this could be due to either loss of viability, due to delay in processing of samples as many time samples from various districts reached the laboratory after 5 days of collection, or due to high contamination rate (4%) or harsh treatment. It is important to maintain balance between culture positivity and contamination by using proper digestion decontamination procedures; 4% contamination is an acceptable norm for MGIT. However, for better yield it’s better to also inoculate the sample on LJ too as contamination is lower on LJ (though isolation rates are lower than MGIT) and separate colonies of MTB and NTM can also be appreciated.
M. fortuitum was the most common species isolated in our study. Similarly Maharashtra also reported M. fortuitum as the predominant species in their study.23 Study from Vellore, Tamil Nadu reported M. chelonae (46%) as predominant species followed by M. fortuitum (41%). 24M. fortuitum was reported in extrapulmonary samples from Lucknow, Uttar Pradesh also,27 whereas M. intracellulare is the predominant species followed by M. abscessus in pulmonary samples in a study from North India.28 The variation in species isolated from different parts of India may be due to the different geographical distribution of NTM. Similarly, from Uganda 63.5% NTM isolates were also M. fortuitum in pulmonary samples.29
Rare NTM species such as M. farcinogenes, M. moriokanese, M. wolinsky, M. goodii, M. peregrinum, and Mycobacterium terrae were isolated in our study by PSC. A study from Delhi22 also reported M. flavescens, Mycobacterium malmoense, Mycobacterium trivale, Mycobacterium triplex, Mycobacteriumsepticum, and M. terrae in different clinical samples. However, a study from Agra, Uttar Pradesh described different NTM species in human infections.30
NTM were found in 1.73% of DRTB suspects and in 0.52% TB suspected samples in our study. Varying positivity has been reported for NTM worldwide in TB suspects: 30.8% from Mexico,31 34.8% from Iran in MDR TB suspects,32 17.6% from Delhi, India,4 and 0.77% from North India in TB suspects.28 These findings highlight the necessity of identification and speciation of NTM so as to initiate appropriate treatment.
In our study, 82.8% of NTM isolates were from pulmonary samples. Similarly, a study from Delhi reported 69.8% NTM in pulmonary and 30.8% in extrapulmonary samples3; 80.9% NTM in pulmonary samples were reported from North India28 and 12.4% NTM in extrapulmonary cases from Delhi,4 whereas Lucknow reported only 29% of NTM in pulmonary samples.33 Another study from Delhi also reported higher NTM in pulmonary (0.28%) than extrapulmonary samples (0.10%).22
Our study shows that prevalence of NTM was higher in males (1.27%) than females (1.03%) as also reported earlier,3,28,33,34 the reasons for this are not well known; some authors have hypothesized that estrogen may have a protective role against NTM,35 while others have reported it to be due to occupational reasons4 however, as we do not have any demographic and socioeconomic details of the patients, it’s difficult to know the reason for same. Patients of a higher age group, above 55 years, were found to have higher positivity for NTM in our study, as reported earlier also3. This could be because older people are more prone to pre-existing lung disease, which may support NTM colonization and infection.
In this study, 20, 17.1, and 14.2% of patients had COPD, bronchial asthma, and bronchiectasis, respectively. These are the main underlying risk factors found to be associated with NTM infections. In a retrospective analysis of NTM patients in China, it was found that NTM infection was more likely accompanied with COPD and arthritis, but not with diabetes, cancer, or bronchiectasis.35 In a study from North India,28 it was also found that 97% patients had bronchiectasis and 32 of these had a history of pulmonary TB.
Among the NTM isolated, surprisingly we found 60% patients had coinfection with MTB; however, significance of such coinfections is yet to be established. A study from United States36 also reported coinfection of TB and NTM but assumed that the patient’s symptoms were largely due to MTB, and role of NTM in disease pathogenesis was not clear. NTM are found more easily in previously damaged or diseased tissue and that could explain why they were isolated from patients with pre-existing TB. Hence, it is important to follow up these patients with coinfection and see response to therapy as the NTM are highly resistant to anti-TB drugs and may be the cause of poor response to therapy. The subculture from MGIT in MTB-positive cases yielded NTM in only 0.30% cases, while PSC yielded 1.19% positivity in coinfection cases. PSC allows growth of only NTM, while LJ allows growth of both MTB and NTM; the low yield on LJ of NTM may occur due to difficulty in separating the nonchromogenic colonies. Some people have tried multiplex PCR from positive MGIT tubes for detection of MTB and NTM that may be clinically more useful as it is a rapid method.37
In our study, five patients had died during treatment; similarly, a study from North India found 10 patients had died of active NTM during follow-up.28
There are some limitations of our study; additional two samples were not available from all the patients with NTM as a result it was not possible to establish pathogenicity in all the patients. Moreover, we had not done multiplex PCR from MGIT growth that could have provided better information on coinfection of MTB and NTM. Another drawback in our study was that subculture on LJ of MGIT-positive cultures was not done immediately but once in 10 to 20 days in batches that may have led to inability to get well-separated colonies of NTM and MTB and missing of NTM. We should have attempted subculture on Middle brook 710 agar plate that may have yielded separate colonies of MTB and NTM in coinfection cases, which we possibly could not identify on LJ as slant is smaller and colonies of MTB and NTM did not separate out properly leading to possibly false low NTM positivity by MGIT.
Conclusion
Prevalence of NTM was very low and is not a cause for great concern. PSC seems to be better than MGIT for isolation of NTM, though we may have missed NTM in MGIT due to poor separation on LJ. To improve the culture isolation rates, reduce contamination and identify coinfection; it will be better to inoculate the sample both on LJ and liquid media. Samples suspected on NTM may also be cultured on PSC even if positive for MTB such that coinfection is not missed. Moreover, inclusion of multiplex PCR may provide rapid results on coinfection of MTB and NTM. It’s important to carry out multicentric studies to know the profile of NTM species causing infections and role of coinfection with NTM in known TB/MDRTB/XDRTB patients with proper follow-up of patients. These being opportunistic pathogen may add to the morbidity and mortality due to MDRB/XDRTB.
Author Contribution
MS conducted the study, wrote the manuscript; BMconceived the study and edited the manuscript; JT performed sequencing experiments and its analysis; and SB planned the experiments and edited the manuscript.
Acknowledgment
We acknowledge the training and support provided by Dr. Partibha Narang and Dr. Rahul Narang from MGIMS Sevagram.
Conflict of Interest
None.
Source of Support
None
Reference
- ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. In: Am J Respir Crit Care Med. Vol 175. 2007. p. :367-416.
- [CrossRef] [PubMed] [Google Scholar]
- Laboratory aspects of clinically significant rapidly growing mycobacteria. Indian J Med Microbiol. 2011;29(04):343-352.
- [CrossRef] [PubMed] [Google Scholar]
- High prevalence of non-tuberculous mycobacterial disease among non-HIV infected individuals in a TB endemic country–experience from a tertiary center in Delhi, India. Pathog Glob Health. 2014;108(02):118-122.
- [CrossRef] [PubMed] [Google Scholar]
- Non-tuberculous mycobacteria in TB-endemic countries: are we neglecting the danger? PLoS Negl Trop Dis. 2010;4(04):e615.
- [CrossRef] [PubMed] [Google Scholar]
- The neglected and often ignored: nontuberculous mycobacteria. J Glob Infect Dis. 2011;3(01):94.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin Chest Med. 2002;23(03):553-567.
- [CrossRef] [PubMed] [Google Scholar]
- A preliminary report on the Mycobacteriosis-like infection among the fish handlers of West Bengal, India. Int J Biol Med Res. 2012;34:2648-2650.
- [Google Scholar]
- Bcnzen. Petroleum Paratfinol un paraffin a!sKohlenstofi” und EngiequellefurMikroben. Zbl Bakt Hyg Abt II. 1913;37:595-609.
- [Google Scholar]
- Paraffin slide culture technique for “baiting” Nontuberculous mycobacteria. Ind J Tub. 2000;47:219-221.
- [Google Scholar]
- Public Health Mycobacteriology: A Guide for the Level III Laboratory. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control; 1985.
- [Google Scholar]
- Manual of Clinical Microbiology. (7th). Washington, D.C.: American Society of Microbiology; 1999. p. :399-437.
- [Google Scholar]
- Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol. 2008;8:48.
- [CrossRef] [PubMed] [Google Scholar]
- Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001;39(10):3637-3648.
- [CrossRef] [PubMed] [Google Scholar]
- Species identification of mycobacteria using rDNA sequencing. Methods Mol Biol. 1998;101:349-361.
- [Google Scholar]
- Spanish Group for Non-Tuberculosis Mycobacteria. Non-tuberculous mycobacteria: patterns of isolation. A multi-country retrospective survey. Int J Tuberc Lung Dis. 2004;8(10):1186-1193.
- [Google Scholar]
- A comparative study for the detection of Mycobacteria by BACTEC MGIT 960, Lowenstein Jensen media and direct AFB smear examination. Indian J Med Microbiol. 2007;25(04):383-386.
- [CrossRef] [PubMed] [Google Scholar]
- GenoType MTBDRplus assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis strains and clinical samples. J Clin Microbiol. 2008;46(11):3660-3667.
- [CrossRef] [PubMed] [Google Scholar]
- Tuberculosis diagnostics in 2015: landscape, priorities, needs, and prospects. J Infect Dis. 2015;211(Suppl. 02):S21-S28.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of an immunochromatographic test for discrimination between Mycobacterium tuberculosis complex & non tuberculous mycobacteria in clinical isolates from extra-pulmonary tuberculosis. Indian J Med Res. 2012;135(06):901-906.
- [Google Scholar]
- Germany: Hain Lifescience GmbH; 2012.
- Evaluation of the Speed-Oligo Mycobacteria assay for the identification of nontuberculous mycobacteria. J Med Microbiol. 2015;64(Pt 3):283-287.
- [CrossRef] [PubMed] [Google Scholar]
- Occurrence of non-tuberculous mycobacterium in clinical samples - a potential pathogen. Ind J Tuber. 2013;xx:71-76.
- [Google Scholar]
- Time to identify and define non-tuberculous mycobacteria in a tuberculosis-endemic region. Int J Tuberc Lung Dis. 2010;14(08):1001-1008.
- [Google Scholar]
- Non tuberculous mycobacteria isolated from clinical specimens at a tertiary care hospital in South India. Indian J Med Microbiol. 2005;23(03):172-175.
- [CrossRef] [PubMed] [Google Scholar]
- The use of paraffin wax metabolism in the speciation of Mycobacterium avium-intracellulare. Tubercle. 1990;71(01):23-28.
- [CrossRef] [PubMed] [Google Scholar]
- Isolation and identification of Mycobacterium tuberculosis with mixed growth from positive MGIT 960 cultures by re-decontamination. J Biotechnol Biomater. 2017;7:267.
- [CrossRef] [Google Scholar]
- Prevalence of nontuberculous mycobacteria among extrapulmonary tuberculosis cases in tertiary care centers in Northern India. Biomed Res Int. 2015;2015 465403
- [CrossRef] [PubMed] [Google Scholar]
- A prospective study of non-tuberculous mycobacterial disease among tuberculosis suspects at a tertiary care centre in north India. Indian J Med Res. 2019;150(05):458-467.
- [CrossRef] [PubMed] [Google Scholar]
- Species and genotypic diversity of non-tuberculous mycobacteria isolated from children investigated for pulmonary tuberculosis in rural Uganda. BMC Infect Dis. 2013;13:88.
- [CrossRef] [PubMed] [Google Scholar]
- Infections due to non-tuberculous mycobacteria (NTM) Indian J Med Res. 2004;120(04):290-304.
- [Google Scholar]
- Nontuberculous mycobacteria in clinical samples with negative acid-fast bacilli. Int J Mycobacteriol. 2017;6(04):391-395.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug resistant tuberculosis versus non-tuberculous mycobacterial infections: a CT-scan challenge. Braz J Infect Dis. 2013;17(02):137-142.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence and species spectrum of both pulmonary and extra-pulmonary non-tuberculous mycobacteria isolate at a tertiary care center. Int J Mycobact 2016:288-293.
- [CrossRef] [PubMed] [Google Scholar]
- Gender susceptibility to mycobacterial infections in patients with non-CF bronchiectasis. Int J Mycobacteriol. 2015;4(02):92-96.
- [CrossRef] [PubMed] [Google Scholar]
- JiS. Retrospective analysis of patients with non-tuberculous mycobacteria from a primary hospital in Southeast China. Sci Rep. 2020;10(01):1060.
- [CrossRef] [PubMed] [Google Scholar]
- Failure to recognize non-tuberculous Mycobacteria leads to misdiagnosis of chronic pulmonary tuberculosis. PLoS One. 2012;5 36902
- [CrossRef] [PubMed] [Google Scholar]
- Mycobacterium tuberculosis and nontuberculosis mycobacteria co-infection: two cases from the sub-Himalayan region of North India in a year. Lung India. 2017;34(05):494-496.
- [CrossRef] [PubMed] [Google Scholar]





