Translate this page into:
Rapid Identification of Yeast Isolates from Clinical Specimens in Critically Ill Trauma ICU Patients
Address for correspondence: Dr. Purva Mathur, E-mail: purvamathur@yahoo.co.in
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Purpose:
The purpose was to evaluate the performance of a commercially available chromogenic Candida speciation media and the Vitek 2 ID system for the identification of medically important yeasts and yeast-like organisms in a routine clinical microbiology laboratory.
Materials and Methods:
A total of 429 non duplicate, consecutive yeast strains were included during the 3.5-year study period. The performance of the Vitek 2 ID system and a chromogenic agar medium was evaluated against the gold standard conventional phenotypic and biochemical identification method for speciation of yeast isolates from trauma patients.
Results:
Candida tropicalis (64%) was the most common Candida species, followed by Candida albicans (14%), Candida rugosa (7%), and Candida parapsilosis (6.5%). Of the 429 isolates, 183 could be identified to species level by all the three methods. Agreement between the chromogenic agar method and conventional methods was 80% for Candida tropicalis, 100% for Candida rugosa, 89% for Candida albicans, and 77% for Candida parapsilosis. Vitek 2 had lower sensitivity, with agreement of 49% for Candida tropicalis, 100% for Candida rugosa, 39% for Candida albicans, and 31% for Candida parapsilosis.
Conclusion:
Thus, in long-term ICU patients, an increasing trend of isolating nonalbicans Candida spp. continues. The chromogenic agar medium is a convenient and economic method to identify commonly isolated species in busy clinical microbiology laboratories.
Keywords
Candida
chromogenic agar medium
identification
trauma
vitek
INTRODUCTION
Despite substantial advances in trauma care, critical traumatic injuries remain one of the leading causes of death in the below 44 years age group.[1] Increased mortality in the critically ill adult trauma population is not solely due to the direct effects of trauma. Patients who survive the initial injury acquire nosocomial infections, which are the leading cause of death in them.[12] Trauma patients usually have prolonged ICU stay and therefore are more prone to acquire hospital-acquired infections (HAIs). These patients are usually treated with broad-spectrum antibiotics, making them more susceptible for Candida infections. In ICUs, the common infections due to Candida sp. are catheter-related blood stream infections (CR-BSIs), intra-abdominal infections, and urinary tract infections (UTIs). Invasive candidiasis is one of the major causes of morbidity and mortality in both immunocompentent and immunocompromised patients, the crude and attributable mortality being high as 40% and 20% respectively.[34] Therefore better management of these infections solely depends on rapid identification and reporting of results.
In the past decade, hospitals across the world have noticed a change in the epidemiology of Candida infections, characterized by a progressive shift from Candida albicans to a predominance of nonalbicans Candida Spp.[4–6] There is growing evidence of the increasing use of azoles causing this epidemiological shift.[47] Characterization to species level helps to identify those strains which might be intrinsically resistant to some of the antifungal agents.
Identification of yeast pathogens by traditional methods requires several days and specific mycological media. These methods are thus labor intensive and time consuming. Several brands of chromogenic media are available for rapid identification of Candida Spp. The chromogenic media contain substrates which react with enzymes secreted by the target microorganisms to yield colonies of varying colors.[8–10]
Commercial automated identification systems have also been developed and are being used in some routine clinical microbiology laboratories. These systems assert for accurate and rapid identification of medically relevant bacteria and yeast.[11–15] The Vitek 2 system (Biomeriux, France) is a fully automated system, dedicated to the identification and susceptibility testing of microorganisms. The ID-YST database of Vitek 2 for yeast identification comprises 51 different taxa, including newly described species, taking into account recent advances in taxonomy.
In this study, we evaluated the performance of a commercially available chromogenic Candida speciation media and the Vitek 2 ID system for the identification of medically important yeasts and yeast like organisms in a routine clinical microbiology laboratory.
MATERIALS AND METHODS
The study was conducted at the Microbiology laboratory of the JPNA Trauma Centre of All India Institute of Medical Sciences (AIIMS) and the Mycology laboratory of the AIIMS hospital. AIIMS is a 2500-bedded, tertiary care, referral, and teaching hospital, where patients are referred from all over India. Its Trauma Centre is a 152-bedded, level 1 Trauma care hospital. The study was conducted from January 2007 to June 2010.
All the consecutive, nonduplicate isolates of yeast recovered from the clinical samples of patients admitted to the trauma center were included in the study.
All the Candida isolates were identified to the species level.
The following strains were taken as controls for evaluation of the various methods: Candida albicans ATCC 10231, Candida tropicalis ATCC 13803, Candida krusei ATCC 14243, and Candida glabarata 15126, Candida kefyr ATCC 204093, Candida guilliermondii ATCC 6260, Cryptococcus noeformans ATCC 14116.
Identification of all clinical isolates was done to the species level by means of conventional mycological methods, chromogenic Candida speciation media, and the Vitek 2 ID system.
For the purpose of comparison, the conventional methods were taken as the gold standard method. The conventional identification was carried out in the Mycology laboratory of AIIMS hospital. All isolates were sent from the Trauma center to that laboratory with a coded number. The identification by other two methods was done at the Microbiology laboratory of the Trauma Centre. Results were compared only when the final report was generated by all the three methods.
Conventional mycological methods
All the candida isolates obtained from clinical specimens were initially subcultured onto sabouraud dextrose agar (SDA) medium and incubated at 37°C till growth appeared. Colonies from the SDA were then plated onto corn meal agar with tween 80 for morphological examination[1617] and triphenyltetrazolium chloride (TTC) medium for determination of pigment production. A germ tube test was performed on yeast-like colonies for presumptive identification of Candida albicans. The strains were further characterized by manual sugar assimilation methods.[1618] The tests were performed according to standard methods and using the above control strains.
Identification on chromogenic agar medium
Growth from SDA slants was plated onto Chrom agar (BD, USA). The plates were incubated at 37°C for 48 hours and the colony morphology (color, size, and texture) were assessed to interpret the identification of species. The interpretation was based on published appearance of various species on chromogenic agar: Parrot green colonies of C. albicans; steel blue colonies of C. tropicalis accompanied by purple pigmentation which diffuses into surrounding agar by growth; larger, fuzzy, rose-colored colonies with white edges of C. krusei; smooth white to light pink colonies of C. glabrata which later became pink; pink to lavender colonies of C. parapsilosis; gray to pale pink of Cryptococcus neoformans and green color with pale borders of C. rugosa [Figure 1]. The quality of the media was checked every time with the ATCC strains. Species were identified when the isolates conformed unequivocally to these morphological features. Two different observers observed the morphology of each isolates. All isolates which gave doubtful morphology or which did not conform to the accepted morphological features were taken as “unidentified” or “misidentified.”

- Colonies grown on CHROMagar Candida for 48 hours at 30°C: (a) C. neoformans, (b) C. albicans, (c) C. tropicalis, (d) C. guilliermondii, (e) C. krusei, (f) C. kefyr, (g) C. glabarata
Vitek 2 ID system
All the strains were subjected to identification by the Vitek 2 system. The tests were performed according to the manufacturer's instructions. Species which were identified as “high discrimination” were taken as the identification of an isolate. The isolates which were “unidentified” or were identified as more than one species (“low discrimination”; for example 50 % probability of identification for three different species) were all taken as “unidentified” or “misidentified” respectively and excluded from analysis.
RESULTS
During the 3.5-year study period, a total of 429 nonduplicate yeast isolates were obtained from the clinical samples of trauma patients. Of these, 238 (55%) were from urine, 174 (40.5%) from blood, 8 (2%) from CVC tips, 4 (1%) from pus, 3 (0.6%) from peritoneal fluid, and 2 (0.4%) from tracheal aspirates. The distribution and sources of Candida species, as identified by the gold standard conventional methods, are shown in Table 1. The ratio of C. albicans to nonalbicans candida isolates was 1: 6.
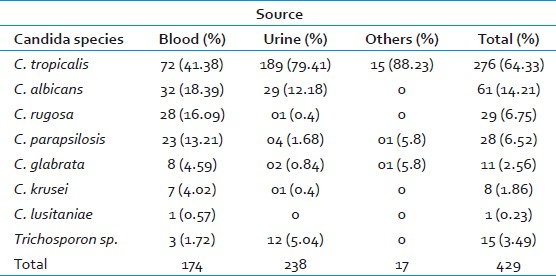
Of these 429 isolates, complete identification to the species level by all the three methods could be obtained in 183 (43%) isolates.
Of these 183 strains, the comparison between chromogenic medium and the gold standard method is shown in Table 2. There was agreement in identification by the chromogenic agar medium in 150 (82%) strains. The performance of the Vitek 2 system against the gold standard is shown in Table 2. Of the 183 strains, only 98 (53%) showed agreement in identification.
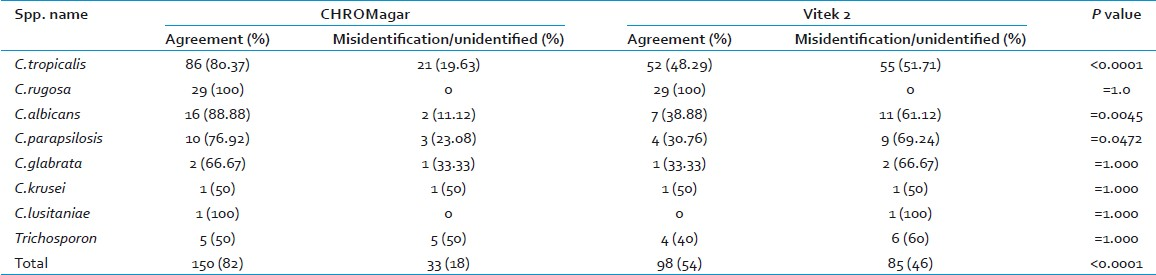
The sensitivity of identification by chromogenic agar for most commonly isolated species was 80% for C. tropicalis, 100% for C. rugosa, 89 % for C. albicans, and 77% for C .parapsilosis (77%) as shown Table 2. Misidentification or nonidentification of C. tropicalis was usually seen due to the difficulty in interpretation of green color, which was misidentified as C. albicans in 11 isolates. In 10 isolates of C. tropicalis, no color was obtained on chromogenic agar medium.
Of the 50 isolates of C. tropicalis misidentified by Vitek 2, 36 were identified as two or three different probable species, 8 were identified with a probability of >90%, but as a different species than conventional methods and 6 had low discrimination of identification. In general, the most common misidentification by Vitek 2 was C. tropicalis or C. albicans identified as Cryptococcus lauretii or Streptomyces ceferrii.
We compared the efficiency of Vitek 2 and chromogenic agar for identification of yeast isolates [Table 2]. Chromogenic agar was certainly the more efficient method over Vitek 2 for identification of most commonly isolated species from our center. Both the methods were apt for identification of some species such as Candida rugosa. But they were not found to be expeditious in identification of less encountered species - Candida krusei, Candida glabarata, Candida lusitaniae.
DISCUSSION
In this study consistent with the published report from different part of the world, Nonalbican Candida species had predominance over C. albicans.[46] C. tropicalis was most common isolate in all samples, followed by C. albicans. A relatively greater proportion of C. tropicalis isolates in our study is concordant with other studies from India.[1920]
We also isolated a large number of C. rugosa from patients admitted to the ICUs, which occurred as a small outbreak.[21] C. rugosa appears to typically produce a readily identifiable and unique color/colony type on chromogenic agar medium. It has been shown in clinical reports and by in vitro testing to be less susceptible to amphotericin B.[2122] Rapid identification of this nonalbican Candida is of great importance to allow provision of appropriate therapy to patients. Also, since it has a tendency to for rapid cross-transmission, early identification is vital.[21]
In our study we evaluated two rapid methods, chromogenic agar medium and fully automated Vitek 2 system for identification of yeast and yeast-like organism. The strains were simultaneously tested by the conventional method. The results indicate that chrom0 agar is able to correctly identify majority of common clinical isolates. The apparent lack of accuracy of Vitek 2 ID in our study is in contrast with previous studies which had shown higher sensitivity.[1423–25]
We observed that for those strains which could not be resolved by chromogenic agar medium alone, sensitivity increased when we used both chromogenic agar and Vitek 2 from 80% to 94% for C. tropicalis and up to 100% for some of the organisms. Thus Vitek 2 ID could be an additional help in those situations where chrom0 agar could not give identification.[26] However there were certain limitations in our study such as there was no isolate of Candida dublinensis wich is difficult to distinguish from Candida albicans on chromogenic agar and very few or no isolates of Candida lusitaniae, Candida krusei, Candida glabarata, and Cryoptococcus neoformans for comparison.
In conclusion, the chromogenic agar medium supported by Vitek 2 ID is a valuable method for identification of commonly encountered, medically important yeast species. The method is easy to interpret and gives rapid results.
ACKNOWLEDGMENTS
We gratefully acknowledge Mrs. Sweety, Mrs. Neelu, Mrs. Raajrani, Mr. Vineet, Mr. Trilok, Mr. Naresh for technical assistance.
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- National Medical Association Surgical Section position paper on violence prevention: A resolution of trauma surgeons caring for victims of violence. JAMA. 1995;73:1788-9.
- [Google Scholar]
- Infection in hospitalized trauma patients: Incidence, risk factors, and complications. J Trauma. 1999;47:923-7.
- [Google Scholar]
- In vitro antifungal susceptibility testing of Candida blood isolates and evaluation of the E-test method. J Microbiol Immunol Infect. 2004;37:335-42.
- [Google Scholar]
- Bench-to-bedside review: Candida infections in the intensive care unit. Crit Care. 2008;12:204.
- [Google Scholar]
- Species distribution and antifungal susceptibility of Candida bloodstream isolates in Kuwait: A 10-year study. J Med Microbiol. 2007;56:255-9.
- [Google Scholar]
- The changing face of candidemia: Emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617-23.
- [Google Scholar]
- Presumptive identification of Candida species other than C. albicans, C. krusei,and C. tropicalis with the chromogenic medium CHROMagar Candida. Ann Clin Microbiol Antimicrob. 2006;5:1.
- [Google Scholar]
- Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J Clin Microbiol. 1996;34:58-61.
- [Google Scholar]
- Use of CHROMagar in the differentiation of common species of Candida. Mycopathologia. 2009;167:47-9.
- [Google Scholar]
- Comparative study of the new colorimetric VITEK 2 yeast identification card versus the older fluorometric card and of CHROMagar Candida as a source medium with the new card. J Clin Microbiol. 2006;44:227-8.
- [Google Scholar]
- Evaluation of six commercial systems for identification of medically important yeasts. Eur J Clin Microbiol Infect Dis. 1998;17:479-88.
- [Google Scholar]
- Evaluation of the AutoMicrobic system for identification of yeasts. J Clin Microbiol. 1982;16:901-4.
- [Google Scholar]
- Comparison of Vitek Yeast Biochemical Card with conventional methods for speciation of Candida. Indian J Path Microbiol. 2000;43:143-5.
- [Google Scholar]
- Comparison of three commercial systems for identification of yeasts commonly isolated in the clinical microbiology laboratory. J Clin Microbiol. 1999;37:1967-70.
- [Google Scholar]
- The yeasts. In: Al - Doory Y, ed. Laboratory Medical Mycology. Philadelphia: Lea & Febiger; 1980. p. :249-83.
- [Google Scholar]
- Medically important fungi. In: A guide to identification (4th ed). WashingtonDC: ASM Press; 2002.
- [Google Scholar]
- Candidiasis and the pathogenic yeasts. In: Wonseiwicz M, ed. Medical Mycology. Philadelphia: W.B. Saunders; 1988. p. :531-81.
- [Google Scholar]
- Candida tropicalis: its prevalence, pathogenicity and increasing resistance to fluconazole. J Med Microbiol. 2010;59:873-80.
- [Google Scholar]
- Emergence of Candida tropicalis as the major cause of fungaemia in India. Mycoses. 2001;44:278-8.
- [Google Scholar]
- Candida rugosa: A possible emerging cause of candidaemia in trauma patients. Infection. 2010;38:387-93.
- [Google Scholar]
- Candida rugosa: A distinctive emerging cause of candidaemia.A case report and review of the literature. Scand J Infect Dis. 2009;41:892-7.
- [Google Scholar]
- Evaluation of the VITEK 2 system for rapid identification of yeasts and yeast-like organisms. J Clin Microbiol. 2000;38:1782-5.
- [Google Scholar]
- A comparison of methods for yeast identification including CHROMagar Candida, vitek system YBC and a traditional biochemical method. Chin Med J. 2001;64:568-74.
- [Google Scholar]
- Evaluation of Vitek 2 for identification of yeasts in the clinical laboratory. Clin Microbiol Infect. 2006;12:591-3.
- [Google Scholar]
- Factors accounting for misidentification of Candida species. J Microbiol Immunol Infect. 2001;34:171-7.
- [Google Scholar]





