Translate this page into:
SARS-CoV-2 Rapid Antigen Detection in Respiratory and Nonrespiratory Specimens in COVID-19 Patients
Address for correspondence: Purva Mathur, MD, Division of Clinical Microbiology, Department of Laboratory Medicine, JPNATC, AIIMS, New Delhi, 110029, India (e-mail: purvamathur@yahoo.co.in).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Rapid antigen testing for coronavirus disease 2019 (COVID-19) available at present provides immediate results at low cost with less expertise and without any need of sophisticated infrastructure. Most of these test kits available are for nasopharyngeal samples. This is a novel study to detect the presence of COVID antigen in samples other than throat and oropharyngeal. Various samples received from patients admitted in the COVID-19 dedicated center were tested for the presence of antigen. Same procedure was followed as done for the nasopharyngeal sample. A total of 150 samples were tested, which included ascitic fluid, pleural fluid, drain fluid, bile, bronchoalveolar lavage, cerebrospinal fluid, endotracheal tube aspirate, sputum, tissue, and urine. Out of 150, 11 (7.33%) were positive and 138 (92.66%) were negative for the antigen test. The COVID-19 antigen test kit, though designed for nasopharyngeal samples, was able to detect the presence of antigen in other clinical samples.
Keywords
COVID-19
SARS-CoV-2
rapid antigen test
nasopharyngeal
nonrespiratory samples
Introduction
A novel human pathogen emerged at the end of 2019 at the city of Wuhan, China and is now known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing the coronavirus disease 2019 (COVID-19).[1] In COVID-19, fever is usually the first symptom with subsequent development of respiratory symptoms.[2,3] Most patients with COVID-19 experience a mild disease course; albeit, approximately 20% develop severe disease with high mortality rate.[2,3]
Laboratory diagnosis and management of COVID-19 has been helpful in combating the spread of SARS-CoV-2. At present, the gold standard for COVID-19 diagnosis is reverse transcription-quantitative polymerase chain reaction (RT-qPCR) which uses nasopharyngeal swabs, throat swabs, or saliva samples.[4] RT-qPCR kits that do not require viral ribonucleic acid (RNA) extraction and high-throughput RT-qPCR systems have also been developed. Although such tests are widely utilized in tertiary care centers and large well-equipped hospitals, they are rarely available in the local clinics which are more approachable for the patients who are under suspicion.
Studies done on rapid antigen tests (RATs) have shown sensitivity of 61.70% and specificity of 98.26% for diagnosis of COVID-19.[5] In the present study, we have used RAT to detect SARS-CoV-2 antigen in samples other than nasopharyngeal.
Material and Methods
This descriptive study was done at a single center dedicated for COVID-19 patients in a tertiary care center in North India. RAT was done on samples like ascitic fluid, pleural fluid, drain fluid, bile, bronchoalveolar lavage (BAL), cerebrospinal fluid, endotracheal tube aspirate (ETA), sputum, tissue, and urine, which were sent to the microbiology laboratory for culture and sensitivity testing. The RAT was done according to the kit insert provided by the STANDARD Q COVID-19 antigen test which is ideally for testing nasopharyngeal sample. Samples were used directly without any dilution. Test was performed as soon as the sample was received and result noted within 30 minutes of the test.
Ethical Approval
Since the samples consisted of routine samples for culture, ethical consent was not taken. However, a blanket ethical clearance for evaluation of all COVID antigen kits was obtained having the ethical committee approval number IEC 668/ July 3, 2020.
Result
A total of 150 patients were included in the study admitted in the hospital. Out of 150 patients, 88 (58.66%) were male and 62 (41.33%) were female. Patients before admission were confirmed as having COVID-19 disease by RT-PCR (73), Truenat Beta (14), Xpert (20), antigen test (17), Xpert and antigen (1), RT-PCR and antigen (6), Truenat and RT-PCR (17), and Xpert and RT-PCR (2). Samples other than nasopharyngeal swab collection were tested. ►Table 1 shows the distribution of sample and their test results. Eleven (7.33%) samples were positive and 138 (92.66%) were negative for the test. Out of the 11 specimens which were positive for the antigen test, 7 were respiratory, 3 were fluid, and 1 was bile. The duration of illness was 6.2 days (95% CI: 3.6–8.7) for the majority of patients with positive specimens. Exceptions were there as for one sample (BAL) test was positive after 2 weeks of onset of symptoms and for another (sputum) patient was asymptomatic. ►Fig. 1 shows the time interval for each positive sample depicting starting of symptoms to the test.
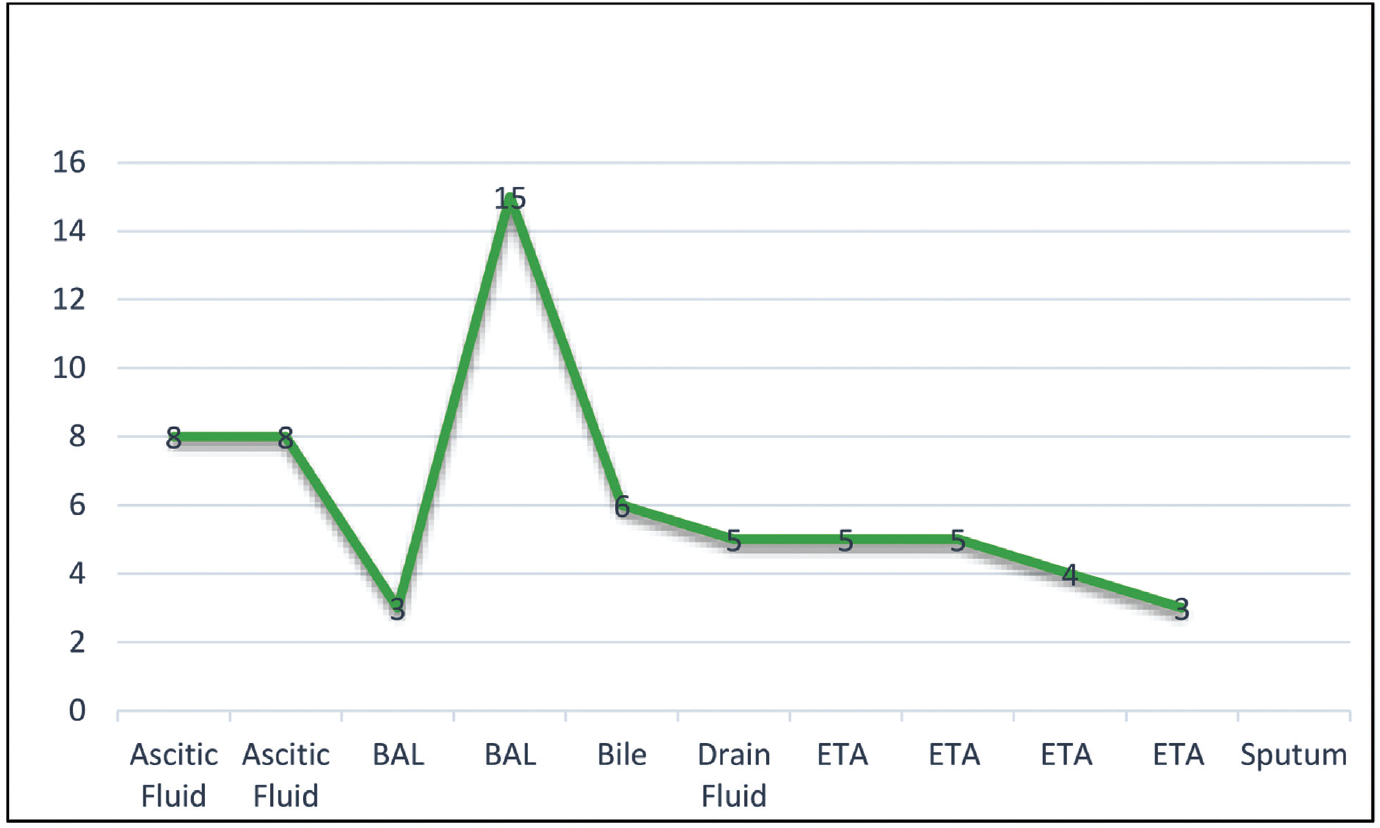
- Time interval from symptoms to the test (days).
| Samples | Antigen test positive/negative | Total |
|---|---|---|
| Ascitic fluid | 2/11 | 13 |
| Drain fluid | 1/6 | 7 |
| Pleural fluid | 0/4 | 4 |
| Cerebrospinal fluid | 0/6 | 6 |
| Bile | 1/3 | 4 |
| Urine | 0/74 | 74 |
| Tissue | 0/1 | 1 |
| Sputum | 1/6 | 7 |
| Endotracheal aspirate | 4/26 | 30 |
| Bronchoalveolar lavage | 2/2 | 4 |
| Total | 11/139 | 150 |
Discussion
Since the outbreak of COVID-19, various strategies are being tried, tested, and followed for the rapid detection, treatment, and containment of this disease. Many have lost their life due to this pandemic. Globally, 101,636,470 have been infected and 2,194,790 have died.[6]
In our country, till now 10,727,240 have suffered and 154,069 have succumbed to this disease.[6] As far as diagnostics is considered, at present rRT-PCR is considered the gold standard. To minimize infrastructure cost, various other modes of nucleic acid detection modalities like Truenat and CBNAAT are also in use. Antigen testing with high specificity[5] has also helped in rapid detection, isolation, and treatment of individuals suspected of having COVID-19. RATs for SARS-CoV-2 are being usually done in throat and nasopharyngeal sample. In a study done in Japan, samples other than nasopharyngeal and throat were also tested with RT-PCR and four different kits of rapid antigen for SARS-CoV-2 were evaluated.[6] Sample tested were gargle lavage, saliva, throat swab, nasal vestibule swab, nasopharyngeal, sputum, and tracheal aspirate. Swabs were transported in BD universal viral transport medium; saliva, sputum, and tracheal aspirate samples were diluted in BD universal viral transport medium if needed and were used as test specimens. Gargle lavages were tested directly. Among the four antigen test kits, one kit was same as what we had used. That kit was able to identify SARS-CoV-2 antigen in saliva (10/27), nasopharyngeal swab (8/18), sputum (1/4), and tracheal aspirate (7/17). In our study, the kit detected the antigen in endotracheal aspirate (4/29), BAL (2/4), sputum (1/7), ascitic fluid (2/13), drain fluid (1/7), and bile (1/4).
With regard to detection of SARS-CoV-2 in nonrespiratory samples, studies have shown its presence in various nonrespiratory samples. RT-PCR has detected SARS-CoV-2 RNA in stool sample in many studies.[7-12] In some studies, where serum or blood were tested, the viral RNA was detected in 30 to 87.5% of patients with COVID-19. The viral RNA shedding was for longer duration in intensive care unit patients (14.63 ± 5.88 days standard deviation [SD]) compared with non-ICU patients (10.17 ± 6.13 days SD).[13-16] This viral RNA has also been detected in ocular tissue too.[17] Ours is a novel study on antigen detection on samples other than nasopharyngeal with positivity of 7.33%. Our study shows that viral antigen and therefore the virus may be present in many other organs apart from the respiratory tract.
Conclusion
The COVID-19 antigen test kit, though dedicated for nasopharyngeal samples was able to detect presence of antigen in other clinical samples. The sample infectivity and transmissibility albeit cannot be assessed with this test alone.
Acknowledgment
The authors would like to thank all participating employees from the JPNATC, AIIMS, Delhi for their help with sample collection, processing, and data collection.
Conflict of Interest
All the authors report no conflict.
Funding
No external funding was received.
References
- China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China,N Engl J Med. 20192020;382(08):727-733.
- [Google Scholar]
- Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506.
- [Google Scholar]
- Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061-1069.
- [CrossRef] [PubMed] [Google Scholar]
- Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249-2251.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of rapid antigen tests for COVID-19. Viruses. 2020;12(12):1420.
- [CrossRef] [PubMed] [Google Scholar]
- Accessed December 13, 2021: https://covid19.who.int. Last updated; January 29, 2021
- Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26(08):1920-1922.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative study on virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Sci China Life Sci. 2020;64(03):486-488.
- [CrossRef] [PubMed] [Google Scholar]
- The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92(07):833-840.
- [CrossRef] [PubMed] [Google Scholar]
- Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(05):434-435.
- [CrossRef] [PubMed] [Google Scholar]
- The time sequences of respiratory and rectal viral shedding in patients with coronavirus disease 2019. Gastroenterology. 2020;159(03):1158-1160.e2.
- [CrossRef] [PubMed] [Google Scholar]
- Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443.
- [CrossRef] [PubMed] [Google Scholar]
- Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl). 2020;133(09):1039-1043.
- [CrossRef] [PubMed] [Google Scholar]
- Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect. 2020;81(01):147-178.
- [CrossRef] [Google Scholar]
- Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(01):386-389.
- [CrossRef] [PubMed] [Google Scholar]
- High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin Infect Dis. 2021;72(09):e291-e295.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of SARS-CoV-2 in human post-mortem ocular tissues. Ocul Surf. 2021;19:322-329. DOI: 10.1016/j.jtos.2020.11.002
- [CrossRef] [PubMed] [Google Scholar]






