Translate this page into:
Simple, rapid, and cost-effective modified Carba NP test for carbapenemase detection among Gram-negative bacteria
Address for correspondence: Dr. Shoorashetty Manohara Rudresh, Department of Microbiology, ESIC Medical College and PGIMSR, Rajajinagar, Bengaluru - 560 010, Karnataka, India. E-mail: rudreshsm@gmail.com
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
PURPOSE:
Detection of carbapenemases among Gram-negative bacteria (GNB) is important for both clinicians and infection control practitioners. The Clinical and Laboratory Standards Institute recommends Carba NP (CNP) as confirmatory test for carbapenemase production. The reagents required for CNP test are costly and hence the test cannot be performed on a routine basis. The present study evaluates modifications of CNP test for rapid detection of carbapenemases among GNB.
MATERIALS AND METHODS:
The GNB were screened for carbapenemase production using CNP, CarbAcineto NP (CANP), and modified CNP (mCNP) test. A multiplex polymerase chain reaction (PCR) was performed on all the carbapenem-resistant bacteria for carbapenemase genes. The results of three phenotypic tests were compared with PCR.
RESULTS:
A total of 765 gram negative bacteria were screened for carbapenem resistance. Carbapenem resistance was found in 144 GNB. The metallo-β-lactamases were most common carbapenemases followed by OXA-48-like enzymes. The CANP test was most sensitive (80.6%) for carbapenemases detection. The mCNP test was 62.1% sensitive for detection of carbapenemases. The mCNP, CNP, and CANP tests were equally sensitive (95%) for detection of NDM enzymes among Enterobacteriaceae. The mCNP test had poor sensitivity for detection of OXA-48-like enzymes.
CONCLUSION:
The mCNP test was rapid, cost-effective, and easily adoptable on routine basis. The early detection of carbapenemases using mCNP test will help in preventing the spread of multidrug-resistant organisms in the hospital settings.
Keywords
Carbapenemases
Carba NP test
CarbAcineto NP test
carbapenem-resistant organisms
metallo-β-lactamase
Introduction
Multidrug resistance is spreading worldwide at an alarming rate among Gram-negative bacteria (GNB). Carbapenems are used as the last line agents to treat infections caused by multidrug-resistant bacteria.[1] The widespread use of carbapenems in clinical practice has led to the development of resistance to these antibiotics. The carbapenem resistance can be due to carbapenemase production, a decrease in bacterial outer membrane permeability with overexpression of AmpC/ESBL or due to overexpression of efflux pump.[12] The genes coding for carbapenemase enzymes are located on bacterial chromosomes or on mobile genetic elements. The horizontal transfer of carbapenemase genes and movement of patients across health-care facilities, countries, and continents has led to the worldwide spread of carbapenem-resistant GNB.[3] Early and rapid detection of carbapenemase-producing GNB helps in the containment of spread of resistance.[34]
Carbapenemase production can be detected by phenotypic and molecular methods. Molecular methods remain as the gold standard for detection of carbapenemases.[5] Although molecular methods are confirmatory, testing may not be immediately available and may be limited by the number of targets detected.[6]
A novel carbapenemase detection test (Carba NP [CNP] test) based on the principle of acidimetry has been developed by Nordmann et al.[1] In the acidimetric method, hydrolysis of beta-lactam ring results in a drop in pH, causing a color change of phenol red indicator from red to yellow. The Clinical and Laboratory Standards Institute (CLSI) with a few modifications recommended the CNP test as a confirmatory test for carbapenemase production.[7] The test requires commercially available bacterial protein extraction reagent (BPER), phenol red indicator, zinc sulfate heptahydrate (ZnSO4 7H2O), and standard grade imipenem powder. The cost of BPER and imipenem standard grade powder is high, leading to increased cost per test. The present study aimed to evaluate modifications of CNP test for the detection of carbapenemases using different inoculum sizes and injectable imipenem-cilastatin powder that resulted in simplified protocols and significant cost reduction per test.
Materials and Methods
Strain collection and source of data
Clinical specimens (blood, urine, sputum, exudates, and fluids) submitted to the microbiology laboratory from September 1, 2015, to December 31, 2015, were studied. Samples were inoculated on 5% sheep blood agar (SBA) and MacConkey agar. All GNB isolated from clinical specimens were used to evaluate the performance of CNP test and its modifications. Bacteria were identified to species level by standard laboratory techniques. The MacConkey agar, 5% SBA, Mueller-Hinton agar, antibiotic discs, phenol red indicator, and chemicals used in the study were procured from HiMedia Laboratories Ltd, Mumbai.
Carba NP test
CNP A solution was prepared by adding phenol red (0.05%) and ZnSO4.7H2O (0.1 mmol/L) to Clinical Laboratory Reagent Water; pH was adjusted to 7.8 ± 0.1, and the solution was stored at 4°C in amber-colored bottles for up to 15 days. The B solution was freshly prepared by adding 12 mg/ml imipenem-cilastatin injectable form (doubling the amount to compensate the cilastatin component; equivalent to 6 mg/ml of imipenem standard grade powder) to A solution and stored at 4°C till it is used. Two calibrated loops (10 μl) of bacterial colony from 18 to 24 h SBA were resuspended in 200 μl of in-house prepared bacterial lysis buffer (Tris-HCL 20 mmol/L and 0.1% Triton X-100) and vortexed for 5 s. Bacterial lysate (100 μl) was added to two microcentrifuge tubes labeled “a” and “b.” Reagents A and B were added to tubes a and b, respectively, incubated at 37°C and readings were taken at 10 min, 30 min, and 120 min by three different observers. The test was considered positive when tube “a” was red and tube “b” was orange/yellow. In a negative test, both tubes remained red. Quality control was achieved using Klebsiella pneumoniae ATCC BAA 1705 (positive control), K. pneumoniae ATCC BAA 1706 (negative control), and plain A and B reagents with lysis buffer (reagent control).
CarbAcineto NP test
Instead of bacterial lysates, 2–3 loops (10 μl) of the test strain were resuspended in 200 μl of 5 M sodium chloride solution and was used as inoculum. The rest of the procedure was similar to CNP test.[8]
Modified Carba NP test
The test strain was grown in peptone water (pH 7) for 2 h, 200 μl of which was used as inoculum. The rest of the procedure was similar to CNP test.
Antibiotic susceptibility testing was done according to CLSI described Kirby-Bauer disc diffusion testing using CLSI recommended antibiotic discs.[7] MIC for imipenem was determined using an agar dilution technique according to CLSI guidelines.[9] The AmpC disc test was done according to Black et al.[10]
All the three tests were run simultaneously on 18–24 h growth cultures of clinical specimens. If scanty growth was observed at 24 h, modified CNP (mCNP) method was performed on the same day, and the other two tests were performed on the next day from sensitivity plates as they required larger inoculum. For carbapenem-resistant isolates, all the three tests were repeated in duplicate before they were subjected for multiplex polymerase chain reaction (PCR). All the isolates were preserved in brain heart infusion broth with 40% glycerol phosphate saline at −70°C for further tests.[11]
Multiplex polymerase chain reaction
All the carbapenem-resistant isolates and ten carbapenem-sensitive isolates were subjected to multiplex PCR using blaNDM, blaIMP, blaVIM, blaOXA-48-like, and blaKPC primers [Table 1]. DNA extraction was done by boiling lysis method,[12] and PCR was done using Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany). Thermal cycling conditions were 95°C for 10 min followed by 30 cycles of 95°C for 1 min, 59°C for 30 s, 72°C for 1 min, and final 72°C for 10 min. Amplification products (20 μl) were analyzed by 1.5% agarose gel electrophoresis in 1x TAE buffer and ethidium bromide with 1 kbp DNA ladder as a size marker.

Sensitivity and specificity
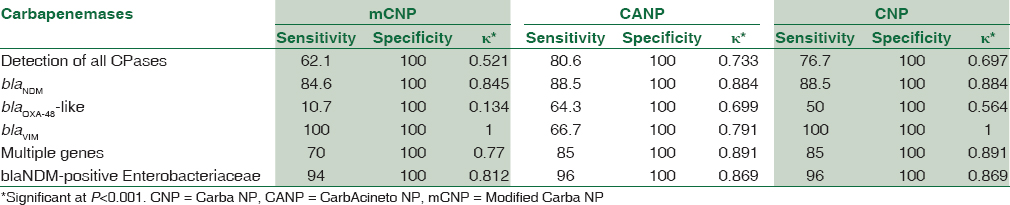
The performance of the three carbapenemase detection methods was evaluated using PCR as the gold standard. The sensitivity (the number of carbapenemase-carrying organisms that were correctly differentiated) and specificity (the number of noncarbapenemase-carrying organisms that were correctly differentiated) of the methods were calculated. Cohen's kappa coefficient was calculated to know the agreement of three tests with the gold standard. The κ >0.65 was considered high agreement and value <0.65 was considered as low agreement between the test and gold standard.
Results
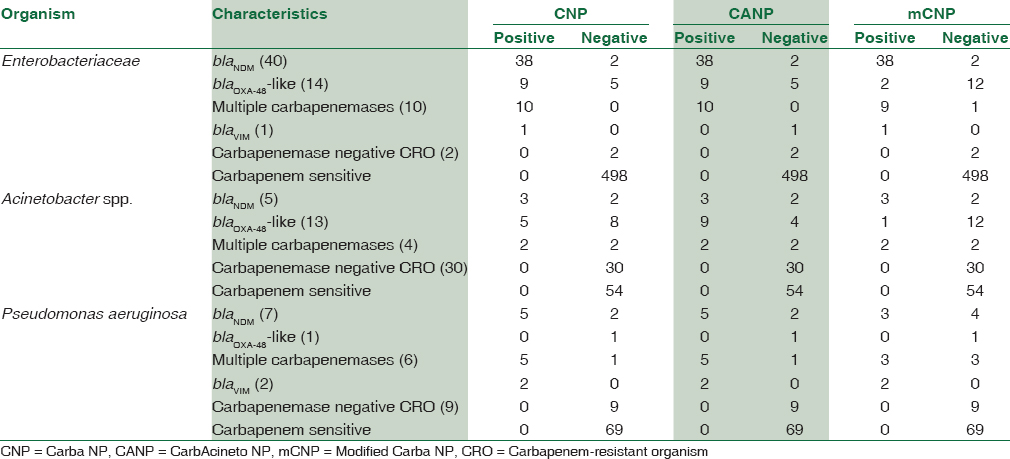
During the study period, a total of 765 gram negative bacteria (Enterobacteriaceae – 565; Acinetobacter spp. – 106; and Pseudomonas aeruginosa – 94) were screened for carbapenem resistance. Carbapenem resistance was found in 144 GNB. The PCR results of all the carbapenem-resistant organisms (CRO) are detailed in Table 2. A total of 103 CRO (71.53%) showed carbapenemase genes by PCR. The PCR test was used as gold standard to compare sensitivity and specificity of the three tests.
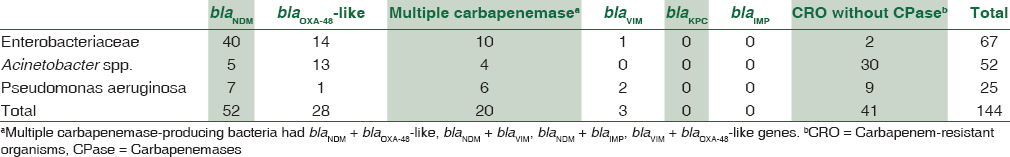
The sensitivity and specificity were 77.7% (95% confidence interval [CI] 68.2%–85%) and 100% (95% CI 99.3%–100%), respectively, for CNP test, 80.6% (95% CI 71.4%–87.5%) and 100% (95% CI 99.3%–100%) for CarbAcineto NP (CANP) test, and 62.1% (95% CI 52%–71%) and 100% (95% CI 99.3%–100%) for mCNP test [Table 3]. The sensitivity of CNP, CANP, and mCNP for carbapenemase-producing Enterobacteriaceae isolates was 89.2%, 87.7%, and 89.2%, respectively [Table 4]. The sensitivity of CNP, CANP, and mCNP for detection of metallo-β-lactamases (MBLs) was 88%, 86.7%, and 81.3%, respectively. Figure 1 shows the results of mCNP for different carbapenemases.
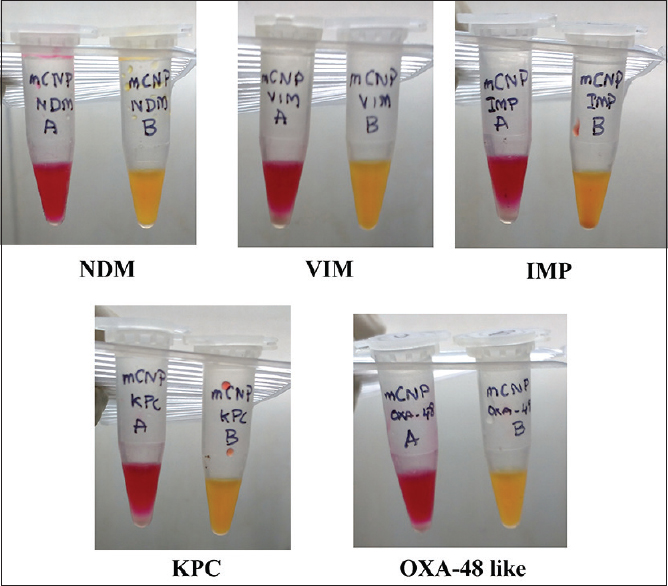
- Performance of modified Carba NP for various carbapenemases
The mCNP had a moderate agreement with PCR for detection of carbapenemases among GNB. The failure of the mCNP test to detect OXA-48-like enzymes resulted in moderate κ value for GNB. The test had good agreement with PCR for blaNDM, blaVIM, and multiple carbapenemase-producing bacteria. Moderate agreement was observed for CNP test for OXA-48-like enzymes detection in comparison to PCR.
There was no interobserver variation in interpretation of results for all the three tests. For MBL producers, positive results were obtained in <10 min by all the three tests. The imipenem MIC for such isolates ranged from 16 to 64 μg/ml. For OXA-48-like enzymes, the test became positive only ≥30 min, and the MIC range for such isolates was 8–32 μg/ml. The isolates with carbapenemase gene, with negative results by all the three tests, showed a MIC range of 0.25–8 μg/ml. All the carbapenem-sensitive isolates remained negative by all the three tests.
Carbapenem-resistant isolates without carbapenemase gene were presumed to be due to other mechanism of resistance like loss of porin channels with the hyperproduction of AmpC or ESBL or overexpression of efflux pump.
Discussion
The CNP test is a colorimetric assay for detection of carbapenem hydrolysis. It was introduced as a confirmatory test for the detection of carbapenemase production among Enterobacteriaceae, Acinetobacter spp., and P. aeruginosa.[7]
The sensitivity and specificity of all the three tests are shown in Table 2. Using CNP test and its two modifications, positive results were obtained consistently for MBL-producing GNB. All the three tests gave inconsistent results for the detection of OXA-48-like carbapenemases. The previous studies showed that CNP test gives false-negative results for few enzymes such as OXA-48, OXA-181, and OXA-244.[45131415] The CNP test uses buffer which counteracts the small amount of acid produced by the weak carbapenem hydrolysis.[8] The use of increased bacterial inoculum and 5M NaCl in CANP test enhanced detection of OXA-48-like enzymes. The low sensitivity of mCNP test for detection of OXA-48-like enzymes can be explained by weak/no lysis of bacteria in peptone water. The lower sensitivity of these tests for OXA-48-like enzymes can be due to weak imipenem hydrolytic activity as evidenced by lower imipenem MIC values and longer time (>30 min) taken for test positivity.[45] Isolates harboring blaOXA-48-like gene with consistent negative results for all the three tests were further studied by quantification of mRNA using real-time reverse transcription PCR (Data not shown). Such strains showed no/low-level expression of blaOXA-48-like genes. Similar finding was made by Dortet et al.[8]
Studies conducted with modifications of CNP test using direct inoculum,[8] injectable imipenem-cilastatin,[513] and different indicators (bromothymol blue)[14] claim equivalent results with CNP test. We tested the Blue-CNP test for carbapenemase detection which showed inconsistent results, and there was interobserver variation in result interpretation (data not shown).
The advantage of mCNP test was, it required very less inoculum compared to CNP and CANP. Clinical specimens in which scanty growth of GNB was seen, the mCNP test was performed on the 1st day and results dispatched within 24 h of sample receipt, while CNP and CANP required 48 h in such conditions. The pH adjustment of peptone water was critical in mCNP test. For consistent results, the pH of peptone water should be adjusted to 7 ± 0.1. The carbapenem-resistant Acinetobacter spp. and P. aeruginosa isolates showing negative mCNP test should be retested by CANP test with heavy inoculum.
The cost of BPER and imipenem standard grade powder used in CLSI described CNP test is high, leading to increased cost per test (4–5$/each test). In the present study, the use of imipenem-cilastatin injectable powder, direct inoculum, and 5M NaCl solution/peptone water instead of BPER reduced the cost to <0.2$/each test. This helped us to screen all GNB isolated from clinical specimens on daily basis. The MBL production contributed to about 50% of carbapenem resistance in our strains. Rapid and consistent detection of MBL by mCNP method helped both the clinicians and the infection control committee to take appropriate actions at the earliest. We could not evaluate the ability of these tests to detect the IMP and KPC type of carbapenemases-producing bacteria as they were infrequently harbored by our isolates. The KPC-producing K. pneumoniae ATCC 1705 consistently showed positive results with all the three tests on repeated testing.
Conclusion
Among the three tests, CANP test performed better than CNP and mCNP test. The mCNP test showed low sensitivity for OXA-48-like enzymes but had a good sensitivity for detection of MBLs. The early detection of carbapenemases using mCNP test contributed in preventing the spread of multidrug-resistant organisms in the hospital settings. The mCNP test is a rapid, consistent, and cost-effective alternative which can be adopted on routine basis in resource-poor countries.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We thank Dr. V. Balaji, Professor and Head, Department of Microbiology, CMC, Vellore, for providing positive control strains for multiplex PCR. We thank Mr. Sridhar, Laboratory Technician, Department of Microbiology, ESIC MC-PGIMSR, Rajajinagar, Bengaluru, for his technical assistance. We thank Dr. Ghanshyam Sharma (PhD), Assistant Professor cum Statistician, Department of Community Medicine, ESIC MC & PGIMSR, Rajajinagar, Bengalore – 10 for statistical assistance.
References
- Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18:1503-7.
- [Google Scholar]
- Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4578-80.
- [Google Scholar]
- Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791-8.
- [Google Scholar]
- Evaluation of the RAPIDEC® CARBA NP, the Rapid CARB Screen® and the carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2015;70:3014-22.
- [Google Scholar]
- Evaluation of the Carba NP test for carbapenemase detection. Antimicrob Agents Chemother. 2014;58:7553-6.
- [Google Scholar]
- Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol. 2013;51:3097-101.
- [Google Scholar]
- M100-S25. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. Wayne, PA: CLSI; 2015.
- CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J Clin Microbiol. 2014;52:2359-64.
- [Google Scholar]
- M7-A9. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically (9th ed). Wayne, PA: CLSI; 2012.
- AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J Clin Microbiol. 2005;43:3110-3.
- [Google Scholar]
- Intergeneric and intrageneric inhibition between strains of Propionibacterium acnes and Micrococcaceae, particularly Staphylococcus epidermidis, isolated from normal skin and acne lesions. J Med Microbiol. 1979;12:71-82.
- [Google Scholar]
- Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J Clin Microbiol. 2011;49:579-85.
- [Google Scholar]
- Simplified protocol for Carba NP test for enhanced detection of carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2015;53:3908-11.
- [Google Scholar]
- Evaluation of the Blue-Carba test for rapid detection of carbapenemases in Gram-negative bacilli. J Clin Microbiol. 2015;53:1996-8.
- [Google Scholar]
- Evaluation of the Rapidec Carba NP test kit for detection of carbapenemase-producing Gram-negative bacteria. Antimicrob Agents Chemother. 2015;59:7870-2.
- [Google Scholar]





