Translate this page into:
Agreement between azithromycin and erythromycin to determine inducible clindamycin-resistant phenotypes in Staphylococcus aureus isolates
*Corresponding author: Nisha Goyal, MBBS, MPH, MD, Department of Microbiology, University College of Medical Sciences & Guru Teg Bahadur Hospital, Dilshad Garden, Delhi 110095, India drnishagoyalucms@gmail.com
How to cite this article: Goyal N, Singh NP. Agreement between azithromycin and erythromycin to determine inducible clindamycin-resistant phenotypes in Staphylococcus aureus isolates. J Lab Physicians. 2024;16:160-3. doi: 10.1055/s-0043-1771244
Abstract
Objectives:
To evaluate agreement in the detection of inducible clindamycin resistance by test method using azithromycin disk with reference method using erythromycin disk and to determine prevalence of inducible clindamycin-resistant phenotypes among Staphylococcus aureus isolates.
Materials and Methods:
A total of 133 non duplicate isolates of S. aureus from clinical samples were included in this prospective study. Agreement between erythromycin (reference method) and azithromycin (test method) disks for detection of inducible clindamycin resistance in S. aureus isolates was assessed.
Statistical Analysis:
Chi-square test was used for analyzing categorical variables (p < 0.05 was considered significant).
Results:
The prevalence of inducible clindamycin resistance (iMLSB) was 15.8%. A 100% agreement was recorded between reference and test methods for detecting inducible clindamycin-resistant phenotypes. For the determination of constitutive resistant phenotypes (cMLSB) among S. aureus isolates, the test method demonstrated an agreement of 94.1%.
Conclusions:
The present study demonstrated an agreement in the identification of inducible clindamycin-resistant phenotypes among S. aureus isolates by both erythromycin and azithromycin disks.
Keywords
inducible clindamycin resistance
iMLSB
constitutive-resistant phenotypes
cMLSB
clindamycin
azithromycin
INTRODUCTION
Staphylococcus aureus is a dominant isolate from the skin and soft tissue infections.[1] It is linked to both health care–associated infections and community-acquired infections.[2] In Staphylococcus spp., the resistance to penicillin and methicillin was first reported in 1944 and 1961 AD, respectively.[3] Methicillin resistant S. aureus (MRSA) is resistant to all the β-lactam antimicrobial agents. With the rising trend of MRSA isolates, the therapeutic options for staphylococcal infections are severely compromised.[4] MLSB family antibiotics are often considered as reserve antibiotics for the treatment of resistant staphylococcal isolates.[1,5] Though antibiotics under the macrolide group such as erythromycin, azithromycin, clarithromycin, and lincosamides (clindamycin and lincomycin) belong to different classes of antimicrobials, they act by the same mechanism of protein synthesis inhibition by binding to 23S rRNA.[6] Clindamycin is a key antimicrobial for the management of staphylococcal infections including both methicillin-susceptible S. aureus (MSSA) and MRSA.[7] Clindamycin is preferred over other macrolides owing to its excellent pharmacokinetic properties including good penetration and distribution into the skin and other soft tissue structures. Furthermore, it has low cost and minimal side effects.[8,9] Unrestrained use of MLSB antibiotics has resulted in an increased incidence of resistance to these antibiotics particularly clindamycin resistance in staphylococcal isolates.[10,11] Bacteria can develop resistance to MLSB antibiotics through several mechanisms including modification of target site, macrolide efflux pump, and enzymatic inactivation of antibiotic.[2]
The erm gene is often responsible for target site modification.[8] Depending on the underlying mechanism for the production of methylase in MLSB, it can be constitutive (cMLSB phenotype) or inducible where methylase production is dependent on the presence of macrolide inducer (iMLSB phenotype).[12,13] Inducible phenotypes are cross-resistant to other group members of the antibiotics including lincosamides and streptogramin B. Thus, the accurate determination of inducible clindamycin resistance is of paramount significance in the management of staphylococcal infections.
Erythromycin (14-membered) and azithromycin (15-membered) macrolides are strong inducers of methylase synthesis. However, spiramycin (16-membered macrolides), clindamycin (lincosamides), and streptogramin B antibiotics are only weak inducers of methylase synthesis. In the presence of the iMLSB phenotype, weaker inducers such as clindamycin may appear active on standard antibiotic susceptibility tests but may lead to therapeutic failure. As per Clinical and Laboratory Standards Institute (CLSI) guidelines, the erythromycin–clindamycin disk approximation test (D test) should be used to determine the presence of inducible clindamycin resistance due to the presence of erm gene in erythromycin-resistant isolates.[14] Azithromycin belongs to the same class of macrolide group of antibiotics. The variation in the basic structure of macrolide resulted in the formation of azalides, a macrolide subclass, that includes azithromycin, clarithromycin, and roxithromycin.[15] Macrolides, 14- or 16-member ring structures, consist of carbon and oxygen, whereas azalides, 15-member ring structures, also contain nitrogen in addition to carbon and oxygen. This variation in the molecule resulted in an enhanced pharmacokinetic property.
In this study, we used the antibiotic discs of azithromycin and erythromycin for the detection of inducible clindamycin resistance and evaluated the agreement in the detection of inducible clindamycin resistance by test method using azithromycin disk with reference method using erythromycin disk. Our secondary objective was to determine the prevalence of inducible clindamycin-resistant phenotypes among S. aureus isolates in our high-volume tertiary care center.
MATERIALS AND METHODS
A prospective study was conducted over a period of 6 months in the bacteriology division of the microbiology department of a tertiary care hospital in Delhi. This study included 133 non duplicate isolates of S. aureus from clinical samples of pus aspirates, body fluids, respiratory tract, and genital tract samples, received in our laboratory for routine bacterial identification and antibiotic susceptibility testing. No separate clinical sample was collected for the purpose of this study.
All clinical samples were inoculated on 5% sheep blood agar, MacConkey agar without crystal violet (HiMedia, India), and incubated at 37°C aerobically for 24 hours. The isolated cream to golden yellow colonies with or without hemolysis on 5% sheep blood agar were further identified by using standard microbiological techniques. The antibiotic susceptibility test was performed using modified Kirby Bauer’s disk diffusion method and interpreted according to CLSI guidelines (CLSI M100, 2022).[14] To detect inducible clindamycin resistance in S. aureus isolates, latest CLSI guidelines were followed; 15 μg erythromycin disk and 2 μg clindamycin disk were spaced 15 to 26 mm apart on Mueller– Hinton agar at 35°C T 2°C, ambient air for 16 to 18 hours.[14]
The detection of inducible clindamycin resistance using the erythromycin disk was considered as the reference test for the same. To determine the agreement between erythromycin (reference method) and azithromycin (test method) disks for inducible clindamycin resistance in S. aureus isolates, azithromycin disk (15 μg) was placed 15 to 26 mm apart from clindamycin disk (2 μg) on the same Mueller–Hinton agar plate on the opposite side of erythromycin disk such that clindamycin disk was placed in the center of the imaginary line joining the erythromycin and azithromycin disks (Figure 1). This ensured the uniformity of distribution of factors that could have influenced the reading of inducible clindamycin resistance using either of the two disks. HiMedia (India) antibiotic disks were used for antibiotic susceptibility testing. Clindamycin resistance was detected as:
Inducible-resistant phenotypes (iMLSB): Resistant to erythromycin and having a clindamycin zone ≤21 mm with a D-shaped zone.
Constitutive-resistant phenotypes (cMLSB): Resistant to both erythromycin and clindamycin.
MS phenotype: Isolates resistant to erythromycin and susceptible to clindamycin without D-zone.
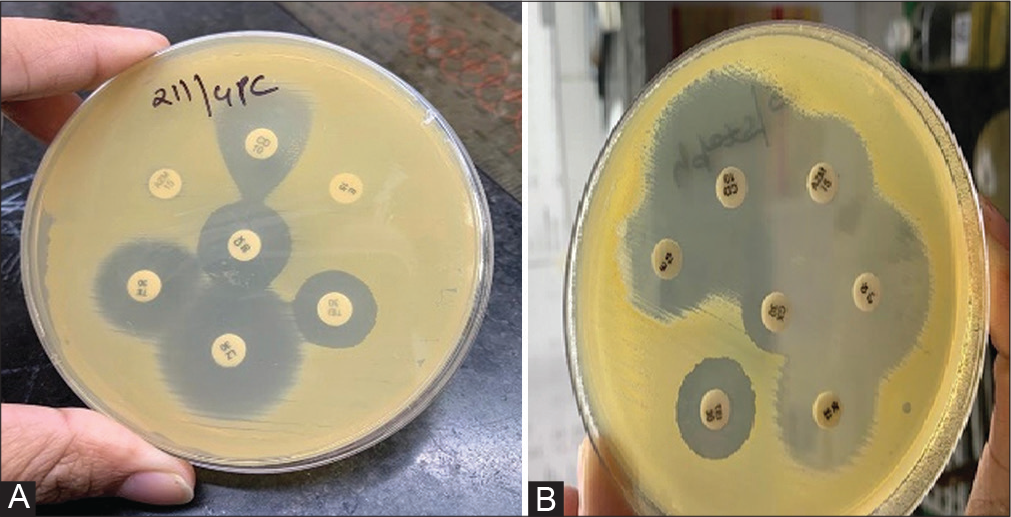
- Inducible-resistant phenotypes (iMLSB) and constitutive-resistant phenotypes (cMLSB) among Staphylococcus aureus isolates as determined by testing clindamycin in combination with erythromycin (reference method) and azithromycin (test method) disks on same Mueller–Hinton agar plate (n ¼ 133). (A) Inducible-resistant phenotypes (iMLSB) and (B) Constitutive-resistant phenotypes (cMLSB) among Staphylococcus aureus isolates as determined by testing clindamycin in combination with erythromycin (reference method) and azithromycin (test method) disks on same Mueller–Hinton agar plate.
For interpretation of inducible clindamycin resistance by azithromycin disk, the CLSI guidelines for interpretation of inducible clindamycin resistance by erythromycin disk were percolated.[14]
Statistical data were analyzed using IBM Statistical Package of Social Sciences (SPSS) software version 20 for Windows software. The chi-square test was used for analyzing categorical variables (p < 0.05 was considered significant).
RESULTS
During the study period, 133 S. aureus isolates were collected prospectively. (Table 1) shows the agreement between the detection of iMLSB, cMLSB, and MSSA phenotypes by testing clindamycin in combination with erythromycin (reference method) and azithromycin (test method) disks. Twenty-one S. aureus isolates that expressed inducible clindamycin resistance (iMLSB) when tested with the reference method also demonstrated the same with the test method. A 100% agreement was recorded between reference and test methods for detecting inducible clindamycin-resistant phenotypes. To determine constitutive resistance phenotypes (cMLSB) among S. aureus isolates, the test method missed one isolate and demonstrated an agreement of 94.1% for the cMLSB phenotype identification. For the determination of susceptibility to individual macrolide antibiotics, 45 S. aureus isolates that were susceptible to erythromycin were also susceptible to azithromycin.
|
Staphylococcus aureus phenotype |
Reference method | Test method |
|---|---|---|
| Inducible-resistant phenotypes (iMLSB) | 21 | 21/21 (100%) |
| Constitutive-resistant phenotypes (cMLSB) |
17 | 16/17 (94.1%) |
| MS phenotype | 50 | 27/50 (54%) |
Figure 2 depicts the distribution of iMLSB, cMLSB, and MS phenotypes among S. aureus isolates as determined by reference and test methods. The prevalence of inducible clindamycin resistance (iMLSB) was 15.8%. Susceptibility to either of the macrolide antibiotic of erythromycin or azithromycin was 33.8%.
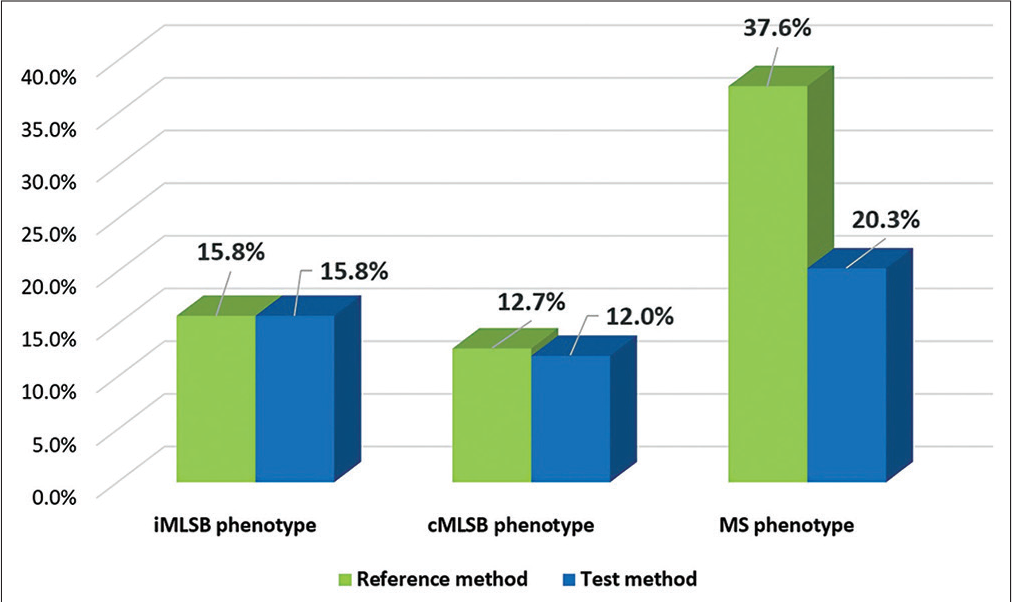
- Distribution of iMLSB, cMLSB, and MS phenotypes among Staphylococcus aureus isolates as determined by reference and test methods (n ¼ 133).
DISCUSSION
Azithromycin, the first azalide, has a more extensive spectrum of activity, better tolerability, and an improved drug interaction profile compared with erythromycin.[15] The addition of nitrogen in azalides makes azithromycin a dibasic molecule (erythromycin is made up of carbon and oxygen only and monobasic in nature) which greatly enhances its pharmacokinetic distribution, sustained release of drugs in the tissues, with their slower elimination rate from our body. Azithromycin has also a unique dosing regimen and postantibiotic effect.[16]
The prevalence of inducible clindamycin resistance varies markedly across different countries and sometimes among hospitals in the same country. The present study reported 15.8% inducible clindamycin resistance among S. aureus isolates. Our findings are in line with the studies.[17,18] In our research, we observed a 100% agreement for the determination of inducible clindamycin resistance between the reference test using erythromycin and the test method using azithromycin. Azap et al in their study also reported a 100% concordance in the detection of inducible clindamycin resistance by azithromycin instead of an erythromycin disk.[19] Furthermore, it is important to mention that azithromycin use has been shown to possess a higher potential for induction of resistance if compared with exposure to other macrolides such as erythromycin and clarithromycin.[8,16]
Resistance against macrolide antibiotics is increasing worldwide. In several severe infections caused by S. aureus such as sepsis, pneumonia, osteomyelitis, and other serious invasive infections, the presence of even a small risk for the emergence of resistance to clindamycin should defer the clinician to its therapeutic use in the management of that particular case. This risk can be easily determined by a positive D-test result.
CONCLUSIONS
The majority of previous research has focused on identifying inducible clindamycin resistance among clinical isolates using erythromycin disk only. To the best of our knowledge, the present study is the first report from India that has demonstrated an agreement in the identification of inducible clindamycin resistance phenotypes among S. aureus isolates by both erythromycin and azithromycin disks.
Authors ’ Statement
The manuscript has been read and approved by all the authors and the requirements for authorship as required have been met, and that each author believes that the manuscript represents honest work.
Authors ’ Contribution
N.G. and N.P.S. contributed to conception and design, acquisition of data, or analysis and interpretation of data; drafting the article; final approval of the version to be published.
Conflict of Interest
None declared.
Source of Funding
None.
References
- Frequency of inducible clindamycin resistance among gram-positive cocci in a tertiary hospital, Tehran, Iran. Iran J Microbiol. 2016;8:243-248.
- [Google Scholar]
- Inducible clindamycin resistance in clinical isolates of staphylococcus aureus in Suez Canal University Hospital, Ismailia, Egypt. J Infect Dev Ctries. 2020;14:1281-1287.
- [CrossRef] [PubMed] [Google Scholar]
- Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu, Nepal. BMC Infect Dis. 2017;17:483.
- [CrossRef] [PubMed] [Google Scholar]
- Methicillin-and inducible clindamycin-resistant Staphylococcus aureus among patients with wound infection attending Arba Minch Hospital, South Ethiopia. Int J Microbiol. 2019;2019:2965490.
- [CrossRef] [PubMed] [Google Scholar]
- The prevalence of inducible and constitutive clindamycin resistance among the nasal isolates of staphylococci. J Clin Diagn Res. 2013;7:1620-1622.
- [CrossRef] [PubMed] [Google Scholar]
- Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43:1716-1721.
- [CrossRef] [PubMed] [Google Scholar]
- Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41:4740-4744.
- [CrossRef] [PubMed] [Google Scholar]
- Macrolide-inducible resistance to clindamycin and the D-test. Pediatr Infect Dis J. 2009;28:1115-1118.
- [CrossRef] [PubMed] [Google Scholar]
- Azithromycin: the first broad-spectrum therapeutic. Eur J Med Chem. 2020;207:112739.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in Gram-positive cocci isolated in a Korean hospital. J Antimicrob Chemother. 2002;49:489-495.
- [CrossRef] [PubMed] [Google Scholar]
- Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J Antimicrob Chemother. 2001;48:315-316.
- [CrossRef] [PubMed] [Google Scholar]
- Inducible clindamycin resistance among clinical isolates of Staphylococcus aureus from Sub Himalayan Region of India. J Clin Diagn Res. 2015;9:DC20-DC23.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482-492.
- [CrossRef] [PubMed] [Google Scholar]
- Performance Standard for Antimicrobial Susceptibility Testing (22nd ed). Wayne, United States: CLSI; 2022.
- [Google Scholar]
- Erythromycin, clarithromycin, and azithromycin: are the differences real? Clin Ther. 1996;18:56-72. discussion 55
- [CrossRef] [PubMed] [Google Scholar]
- Blood, tissue, and intracellular concentrations of azithromycin during and after end of therapy. Antimicrob Agents Chemother. 2013;57:1736-1742.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of inducible and constitutive clindamycin resistance among Staphylococcus aureus isolates in a tertiary care hospital, Eastern India. Avicenna J Med. 2016;6:75-80.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of inducible clindamycin resistance in Staphylococcus aureus at a tertiary care hospital: implications for clinical therapy. Int J Curr Microbiol Appl Sci. 2014;3:720-725.
- [Google Scholar]
- Incidence of inducible clindamycin resistance in staphylococci: first results from Turkey. Clin Microbiol Infect. 2005;11:582-584.
- [CrossRef] [PubMed] [Google Scholar]





