Translate this page into:
Multiplex PCR assay for simultaneous detection and differentiation of Entamoeba histolytica, Giardia lamblia, and Salmonella spp. in the municipality-supplied drinking water
Address for correspondence: Prof. Sarman Singh, All India Institute of Medical Sciences, Ansari Nagar, New Delhi - 110 029, India. E-mail: sarman.singh@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
BACKGROUND:
The contamination with Entamoeba histolytica, Giardia lamblia, and Salmonella spp. in drinking water is the most prevalent in Indian subcontinent, but often difficult to detect all these pathogens from the drinking water.
MATERIALS AND METHODS:
A multiplex polymerase chain reaction (mPCR) method was developed to detect contamination of municipality-supplied drinking water with E. histolytica, G. lamblia, and Salmonella spp. The primers were designed to target small subunit of 16S rRNA type gene of E. histolytica and G. lamblia, and invasive A gene of Salmonella typhimurium. The optimized mPCR assay was applied on 158 municipality-supplied drinking water samples collected from Delhi.
RESULTS:
Out of total 158 water samples, 89 (56.32%) were found positive for the targeted pathogens by mPCR while conventional methods could be detected only in 11 (6.96%) samples. The mPCR assay showed 100% sensitivity and specificity for these pathogens in comparison with culture and microscopic detection. Of the 89 mPCR-positive samples, G. lamblia, E. histolytica, and Salmonella spp. were present in 35 (22.15%), 26 (16.45%), and 28 (17.72%), respectively. Nine (5.69%) samples were positive for both E. histolytica and G. lamblia, 10 (6.32%) were positive for G. lamblia and Salmonella spp., and 8 (5.06%) had Salmonella spp. and E. histolytica. Nonetheless, 3 (1.89%) samples were positive for all three pathogens.
CONCLUSIONS:
The present assay is an alternative to conventional methods to serve as highly sensitive, specific, and economical means for water quality surveillance to detect the outbreak caused by E. histolytica, G. lamblia, and Salmonella spp. pathogens.
Keywords
Drinking water
Entamoeba histolytica
Giardia lamblia
multiplex polymerase chain reaction
Salmonella species
Introduction
Over the last 100 years, provision of the safe and pure drinking water is one of the most important advances for public health regulatory bodies. However, still >1 billion people do not have the access of pure water and >2.4 billion population lacks adequate sanitation. The unhygienic conditions and contaminated water accounts for >1 billion cases of waterborne infection annually and contributing about 10.25 million deaths worldwide, 95% of the diseases are in children of <5 years of age.[1] The etiological agents for these infections are viruses (Norwalk-like viruses, rotaviruses, and enteric adenoviruses), bacteria (Campylobacter jejuni, Shigella, Salmonella, enterotoxigenic Escherichia coli, and cytotoxigenic Clostridium difficile), and parasites (Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum).[234] The presence of E. coli and free-living amoeba indicates highly sensitive indicator of fecal contamination and signifies the presence of other pathogenic bacteria, virus, and protozoa in drinking water.[5] The presence of fecal contaminants like E. histolytica, G. lamblia, and Salmonella spp. in drinking water is important because these are most common causative agent of gastrointestinal illness to the humans.
E. histolytica is causative organism for amoebic liver abscess disease. Worldwide, the disease manifestation causes 100,000 deaths annually.[6] The presence of E. histolytica is found in >90% of the water-borne outbreaks caused by pathogenic protozoan and rest 10% are associated with the food.[7]
G. lamblia is also common intestinal pathogenic organism and has been associated with numerous outbreaks of water-borne diseases.[8910] It is endemic in many countries, predominantly in the third world, with 30%–50% of children harboring parasite continuously or intermittently.[11] Giardia duodenalis synonym of G. intestinalis and G. lamblia is found in wide range of mammalian hosts.[12] Presence of 5 or more Giardia cyst in 100 liters of drinking water is sufficient to cause Giardia sis disease.[13] Giardia is the common cause of parasitic gastroenteritis in human and is a major health problem worldwide.[14] Giardia survives for months in water without nutrients and exists as a hardy, highly infectious cyst, and oral ingestion causes the infection. The common symptoms persist for 2–6 weeks, which includes diarrheal, nausea, stomach cramps with gas, poor nutrient absorption, and weight loss.
Salmonella is Gram-negative, rod-shaped, and nonspore-forming Enterobacteriaceae.[15] It is important bacterial pathogen implicated in water and food-borne outbreaks.[16] Based on lipopolysaccharide (O), flagellar protein (H) and sometimes, the capsular (Vi) antigen, the family is classified into serovars (serotypes) and based on virulence factors. The Salmonella spp. is responsible for typhoid fever to 16 million persons, gastroenteritis illness to 1.3 billion individual and causes 3 million deaths yearly worldwide.[17] Microscopic observation, culture growth on specific media, and biochemical tests are the used methods for detection of these in water. However, these methods are labor-intensive, time-consuming and costly. Therefore, the use of molecular techniques, i.e., polymerase chain reaction (PCR) is an alternate choice. PCR assay can detect the small quantity of DNA of pathogens present in the water and waste-water samples. As it is considered the better tool and has a potential to overcome many of the limitations posed by conventional methods. It will be more economical and practical if assay detects the multiple pathogenic species in a single PCR reaction tube. The use of multiplex PCR (mPCR) for pathogen detection in clinical and environmental samples is well known.[1819] The mPCR provides a smart choice against monoplex PCR. Therefore, the purpose of the present study was to develop and evaluate the mPCR assay to detect the prevalence of two different etiological agents, i.e., parasite and bacteria particularly E. histolytica, G. lamblia, and Salmonella spp. in drinking water which is a novelty of this study.
Materials and Methods
Study Site and Consent
This study was carried out in Division of Clinical Microbiology and Molecular Medicine, Department of Laboratory Medicine, All India Institute of Medical Sciences, New Delhi, India. Written consents duly signed were taken from the owner of the house or sample collection site before collecting the samples.
Assay controls
The DNA of E. histolytica (HM-1 strain) and G. lamblia (Portland-I strain) and Salmonella spp. were used as a control for mPCR. The E. histolytica and G. lamblia DNA was kind gift of Dr. Alok Bhattacharya, Jawaharlal Nehru University, New Delhi, and Dr. Rakesh Sehgal, Post Graduate Institute of Medical Education and Research, Chandigarh, India. For Salmonella spp. DNA, a single typical colony was picked from known culture agar plate and inoculated in 5 ml of nutrient broth and was incubated for overnight at 37°C. The bacterial cells harvested by centrifuging at 6000 x g for 10 min at 4°C. DNA was isolated by phenol–chloroform extraction method,[20] and isolated DNA was dissolved in 30 μL of Tris-EDTA (TE) buffer.
Design of primers
The primers were designed by using primer-BLAST online tool at NCBI to target small ribosomal subunit 16S RNA gene of E. histolytica (accession no. X64142), G. lamblia (accession no. M54878), and invasive A gene of Salmonella typhimurium (accession no. U3237). The primers were custom synthesized by MWG Biotech Private Limited, Bengaluru, India [Table 1].
| Primer | Sequence (5’- 3’) | Oligomers | Targeted gene | Amplicon (bp) |
|---|---|---|---|---|
| E. histolytica | ||||
| E-F | AGG TTT CAG TCT CGT TCG TTA C | 22 | Small subunit 16S rRNA gene | 477 |
| E-R | ATC CAT GAT CGC TAT AAG ATG CAC | 24 | ||
| G. lamblia | ||||
| G-F | GTC CAA CCG TTG TCC TGA G | 19 | Small subunit 16S rRNA gene | 74 |
| G-R | ACG CTC TCC CCA AGG AC | 17 | ||
| Salmonella spp. | ||||
| S-F | GTT GTC TTC TCT ATT GTC ACC GT | 23 | Invasive-A gene | 293 |
| S-R | CGA AAT ACC GCC AAT AAA GTT CAG | 24 |
E. histolytica=Entamoeba histolytica, G. lamblia=Giardia lamblia
Standardization of multiplex polymerase chain reaction assay
The DNA of all three pathogens was used for mPCR standardization. The 3 μL DNA, (100 ng) of each pathogen was spiked in 10 μL of nuclease free water to serve as positive control, and nontargeted enteric pathogens' DNA was used as negative control. The PCR reaction mixture was prepared by adding 3 μL (100 ng) of template DNA, 2.5 μL Taq polymerase buffer with 1.5 mM MgCl2 (×10) (Fermentas, Canada), 0.5 μL dNTPs (10 mM) (Fermentas, Canada), 1.5 μL forward and reverse primers each (10 pmol/μL), 0.5 μL Taq DNA polymerase (3U/μL) (Fermentas, Canada); final volume was made up to 25 μL with nuclease-free sterile water.
The reaction was carried at temperature of 95°C for 5 min before initial PCR amplification cycle. The temperatures cycle used for amplification were 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s for 35 cycles followed by final extension at 72°C for 7 min in thermal cycler (Bioer's XP thermal cycler). The amplified gene products were resolved in 1.8% agarose gel.
Evaluation of multiplex polymerase chain reaction assay on municipality-supplied drinking water samples
Collection, filtration, and concentration of water
A total of 158 municipality-supplied drinking water samples (20 L volume) were collected from different residential area of Delhi between periods of April, 2007 and May, 2010 in 25 L polypropylene container and carried to laboratory in minimal period of time. The samples were filtered and concentrated through 0.22 μm polyethersulfone pellicon-2 minifilter (Millipore Corporation, Billerica, MA01821) in a masterflex pellicon-2 miniholder using a Millipore vacuum pressure pump at 115V and 3 ψ. The filtration and concentration unit was rinsed with double-distilled water followed by 0.1N NaOH before and after each use to prevent cross contamination between two samples. Finally, 20–25 ml of sample was further centrifuged at 12,000 rpm for 20 min to concentrate it up to 0.5 ml volume.
DNA isolation from the water retentate for Entamoeba histolytica, Giardia lamblia, and Salmonella spp.
From the concentrated water retentate, 200 μL volume was used for E. histolytica, G. lamblia and Salmonella spp. DNA isolation. The DNA extraction was carried out by QIAmini stool DNA extraction kit (Qiagen, USA) as per the manufacturer's instructions. The extracted DNA was used for the presence of pathogen detection by standardized mPCR assay. The amplified gene products specific to targeted nucleotide sequences were visualized by agarose gel electrophoresis.
Microscopy and culture methods for Entamoeba histolytica, Giardia lamblia, and Salmonella spp detection
The presence of targeted pathogens in concentrated water retentate was detected by direct microscopy and culture techniques. The E. histolytica G. lamblia parasites (ova and cysts) detected by microscopically and Salmonella spp. by culture techniques followed by biochemical methods. Lugol's iodine and direct saline wet mount of concentrated water was used to visualize cyst and trophozoites of G. lamblia and E. histolytica. For Salmonella spp., a loopful retentate was streaked on MacConkey agar plate and was incubated overnight at 37°C for 18–24 h. The presence of typical single colony obtained was subculture for biochemical analysis and agglutination assay with Salmonella specific anti-sera (Serobac, BioRad) for confirmation.
Results
Optimization of the multiplex polymerase chain reaction assay
The mPCR for simultaneous detection of E. histolytica, G. lamblia, and Salmonella spp. were standardized with DNA of control strains. Developed mPCR assay precisely amplified the targeted genes of E. histolytica, G. lamblia, and S. typhimurium as 477 bp, 74 bp and 293 bp respectively [Figure 1]. The mPCR was evaluated with DNA of other nontargeted enteric pathogens such as E. coli, Iodamoeba buestchili, and Endolimax nana. No amplification was seen in the spiked or in the neat specimens (data not shown), giving us 100% specificity.

- Standardization of multiplex polymerase chain reaction assay; Lane 1: DNA ladder 50 bp; Lane 2: Entamoeba histolytica (477 bp); Lane 3: Salmonella typhimurium (293 bp); Lane 4: Giardia lamblia (74 bp); Lane 5: Positive control; Entamoeba histolytica + Salmonella typhimurium + Giardia lamblia; Lane 6: Negative control DNA
Specificity and sensitivity of multiplex polymerase chain reaction assays primer pairs
The primer specificity was tested with DNA of targeted control pathogens and nontargeted but related organisms, i.e., E. coli, I. buestchili, and E. nana. There was no amplification in the DNA of nontargeted pathogens while desired amplicons were seen in all targeted pathogens. A total of ten amplified products were randomly selected and sequenced. The assembled sequences were compared with reference sequences by BLAST analysis, which showed 98%–100% homology indicating high specificity of our mPCR. The minimum concentration detectable by species-specific PCR varied in sensitivity according to the species. As determined by agarose gel electrophoresis, the optimal sensitivity for detection of E. histolytica, G. lamblia, and Salmonella spp. were 0.0651 ng/μL, 0.0138 ng/μL, and 0.0762 ng/μL, respectively [Figure 2a-c].
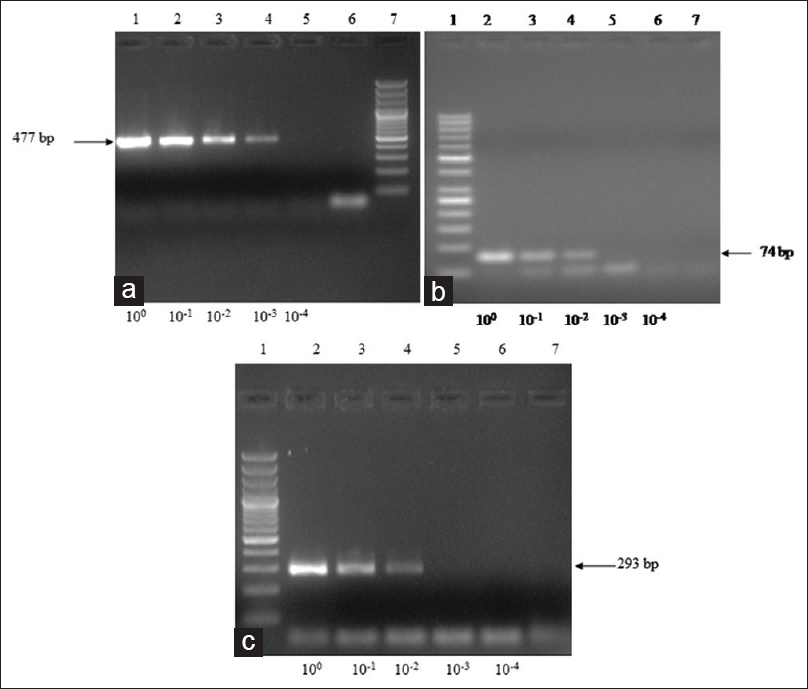
- Sensitivity analysis of multiplex polymerase chain reaction on the DNA isolated from Entamoeba histolytica, Giardia lamblia, and Salmonella typhimurium. The DNA concentration was measured by spectrophotometer at 260 nm. (a) Amplification from serial four-fold dilution of Entamoeba histolytica DNA; Lane 1: 100; 2: 10-1; 3: 10-2; 4: 10-3; 5: 10-4; 6: Negative DNA; 7: DNA ladder-50 bp. (b) Amplification from serial four-fold dilution of Giardia lamblia DNA; Lane 1: DNA ladder-50 bp; 2: 100; 3: 10-1; 4: 10-2; 5: 10-3; 6: 10-4; 7: Negative control DNA. (c) Amplification from serial four-fold dilution of Salmonella typhimurium DNA; Lane 1: DNA ladder-50 bp; 2: 100; 3: 10-1; 4: 10-2; 5: 10-3; 6: 10-4 dilution; 7: Negative control DNA
Detection of Entamoeba histolytica, Giardia lamblia, and Salmonella spp. by multiplex polymerase chain reaction
The mPCR-based detection of E. histolytica, G. lamblia, and Salmonella spp. in 158 municipality-supplied drinking water samples was significant finding of the study. These samples included 05 (3.16%) samples microscopically positive for G. lamblia, 02 (1.26%) for E. histolytica and 05 (3.16%) were culture positive for Salmonella species. One sample (0.63%) was positive for G. lamblia and E. histolytica; but, none of these samples were found positive for Salmonella spp. and E. histolytica. The positivity rate of in-house mPCR was significantly high as compared to standard methods [Table 2]. From total samples, 89 (56.32%) were found positive. Of these, 35 (22.15%) were positive for G. lamblia, 26 (16.45%) for E. histolytica, and 28 (17.72%) for Salmonella spp. [Table 3]. There were 9 (5.69%) samples positive for both E. histolytica and G. lamblia whereas 10 (6.32%) were positive for G. lamblia and Salmonella spp. and 08 (5.06%) for Salmonella spp. and E. histolytica. Only 03 (1.89%) samples were positive for all three pathogens [Figure 3].
| Conventional method | mPCR assay | |||||
|---|---|---|---|---|---|---|
| Microscopy | Culture | |||||
| E. histolytica | G. lamblia | Salmonella spp. | Salmonella spp. | G. lamblia | E. histolytica | |
| Positive (%) | 2 (1.26) | 5 (3.16) | 4 (2.53) | 28 (17.72) | 35 (22.15) | 26 (16.45) |
| Negative (%) | 156 (98.73) | 153 (96.83) | 154 (97.46) | 130 (82.27) | 123 (77.84) | 132 (83.54) |
| Total (%) | 158 (100) | |||||
E. histolytica=Entamoeba histolytica, G. lamblia=Giardia lamblia, mPCR=Multiplex PCR, PCR=Polymerase chain reaction
| Targeted pathogens (alone or in combination) | By standard method (%) | By mPCR assay (%) |
|---|---|---|
| E. histolytica only | 2 (1.26%) | 26 (16.45%) |
| G. lamblia only | 5 (3.16%) | 35 (22.15%) |
| Salmonella spp. only | 4 (2.53%) | 28 (17.72%) |
| E. histolytica and G. lamblia both | 1 (0.63%) | 9 (5.69%) |
| G. lamblia and Salmonella spp. both | 1 (0.63%) | 10 (6.32%) |
| E. histolytica and Salmonella spp. both | 0 (0.0%) | 8 (5.06%) |
| E. histolytica, G. lamblia and Salmonella spp. all three pathogen | 0 (0.0%) | 3 (1.89%) |
E. histolytica=Entamoeba histolytica, G. lamblia=Giardia lamblia, mPCR=Multiplex PCR, PCR=Polymerase chain reaction
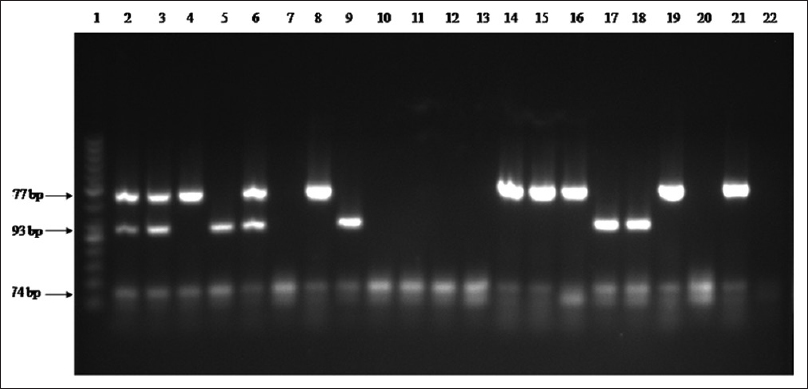
- Multiplex polymerase chain reaction amplification of Entamoeba histolytica, Giardia lamblia and Salmonella spp. in collected water. Lane 1: DNA ladder-50 bp; Lane 2: Positive control (Entamoeba histolytica + Salmonella typhimurium + Giardia lamblia); Lane 3–21: Water samples; Lane 22: Negative control DNA
Discussion
The mPCR assays are technically more superior to conventional methods such as microscopic and culture technique due to their accuracy and reliability. PCR-based detection of different water-borne pathogen in environment samples has been done, and it can be applied as promising substitute for the existing conventional methods.[212223] Although the monoplex PCR is well accepted for initial standardization, it becomes cumbersome and costly for identifying multiple pathogens in the tested samples. Because in the monoplex PCR, the laboratory has to perform the test individually for each suspected pathogen. In this process, very often, the copathogens are missed. However, mPCR can screen numerous microbial organisms simultaneously in a single tube setup. Desouky et al., (2003) developed an mPCR assay to monitor water quality for pathogenic bacteria presence.[24] Whereas Rai et al.[25] used immunofluorescence assay (IFA) combined with polymerase chain reaction for the detection of microbial load mainly Giardia, Entamoeba, and Cryptosporidium.[25] The mPCR assays have been developed against enteric pathogens also but none of these assays detect parasites (G. lamblia and E. histolytica) and bacteria (Salmonella species) simultaneously in this combination. These pathogens are most commonly found in contaminated water and are responsible for various gastro illness of human. The detection of these pathogens by PCR is more sensitive than microscopic method[2627] and is also shown in our result. Out of 158 samples, only 11 (6.96%) samples were positive by conventional techniques while 89 (56.32%) samples were positive with mPCR.
The control of water-borne outbreak needs efficient and sensitive detection of disease causing pathogens. However, in spite of the various advantages of the mPCR assay, the initial standardization requires skill manpower and good laboratory setup. Furthermore, even after preliminary evaluation, depending on the nature of sampling, it can also be affected by inhibitory substances being present in the environmental samples. This can impact the sensitivity of the assay. In the present study, the inhibitors present in the municipality-supplied drinking water were removed by use of inhibit-X tablets supplied along with the DNA extraction kit. Therefore, the scope of our finding provides a new detection method for easy surveillance of such pathogens.
Conclusions
The mPCR assay is highly sensitive and specific in comparison to conventional microscopic and culture techniques. The developed novel assay is suitable for routine screening of drinking water and ruling out the contamination with E. histolytica, G. lamblia, and Salmonella spp. All three pathogens can be detected in one reaction and in a single reaction tube.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The financial support from the Department of Biotechnology, Government of India (BT/PR-8172/BCE/08/482/2006) is acknowledged. We are thankful to Prof. A Bhattacharya of School of Life Sciences, Jawaharlal Nehru University New Delhi India and Dr. R Sehgal, PGIMER, Chandigarh for providing E. histolytica and G. lamblia DNA. Further, we would like to extend our thanks to Ms. Veena Balooni and Ms. Shalini Singhal for their technical assistance in bacteriological identification.
References
- Interventions to improve water quality for preventing diarrhoea. Cochrane Database Syst Rev. 2006;(3):CD004794.
- [Google Scholar]
- Diarrhea in developed and developing countries: Magnitude, special settings, and etiologies. Rev Infect Dis. 1990;12(Suppl 1):S41-50.
- [Google Scholar]
- Giardia sis as a re-emerging infectious disease and its zoonotic potential. Int J Parasitol. 2000;30:1259-67.
- [Google Scholar]
- Seasonal prevalence of intestinal parasites in the United States during 2000. Am J Trop Med Hyg. 2002;66:799-803.
- [Google Scholar]
- Biological contamination of drinking water in New Delhi. Isolation of free living amoebas as a highly sensitive index of water contamination. Water Int. 2000;25:403-9.
- [Google Scholar]
- Giardia, Cryptosporidium, and Cyclospora and their impact on foods: A review. J Food Prot. 1999;62:1059-70.
- [Google Scholar]
- A polymerase chain reaction procedure for the detection of Salmonella spp. Within 24 hours. J Food Prot. 1998;61:1039-42.
- [Google Scholar]
- Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77:487-8.
- [Google Scholar]
- water-borne Giardia cysts and Cryptosporidium oocysts in the Yukon, Canada. Appl Environ Microbiol. 1993;59:67-73.
- [Google Scholar]
- Detection of Giardia in environmental waters by immuno-PCR amplification methods. Curr Microbiol. 1998;36:107-13.
- [Google Scholar]
- Immune response to Giardia lamblia in a water-borne outbreak of Giardiasis in Sweden. J Med Microbiol. 1992;36:347-52.
- [Google Scholar]
- Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol Today. 2000;16:210-3.
- [Google Scholar]
- Application of monitoring data for Giardia and Cryptosporidium to boil water advisories. Risk Anal. 2001;21:1077-85.
- [Google Scholar]
- Detection of pathogenic bacteria in shellfish using multiplex PCR followed by CovaLink NH microwell plate sandwich hybridization. J Microbiol Methods. 2003;53:199-209.
- [Google Scholar]
- Food-borne Microbial Pathogens: Mechanisms and Pathogenesis. New York: Springer: Science; 2008.
- Multiplex PCR protocol for the diagnosis of staphylococcal infection. J Clin Microbiol. 2001;39:3332-8.
- [Google Scholar]
- Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40:3613-9.
- [Google Scholar]
- Molecular Cloning: A Laboratory Manual. New York: CSHL Press; 1989.
- A PCR-based method for monitoring Legionella pneumophila in water samples detects viable but noncultivable legionellae that can recover their cultivability. Appl Environ Microbiol. 2008;74:4817-24.
- [Google Scholar]
- Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: Utility for daily practice. Int J Hyg Environ Health. 2008;211:403-11.
- [Google Scholar]
- Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Appl Environ Microbiol. 2009;75:2798-801.
- [Google Scholar]
- Multiplex-PCR and PCR-RFLP assays to monitor water quality against pathogenic bacteria. J Environ Monit. 2003;5:865-70.
- [Google Scholar]
- Detection of Giardia entamoeba and Cryptosporidium in unprocessed food items from Northern India. World J Microbiol Biotechnol. 2008;24:2879-87.
- [Google Scholar]
- Detection of Cryptosporidium parvum oocysts in faeces: Comparison of conventional coproscopical methods and the polymerase chain reaction. Vet Parasitol. 1996;61:5-13.
- [Google Scholar]
- Real-time PCR for the detection of Giardia lamblia. Mol Cell Probes. 2003;17:223-5.
- [Google Scholar]





