Translate this page into:
Neutrophil-to-Lymphocyte Ratio and Platelet-to-Lymphocyte Ratio and Their Role as Predictors of Disease Severity of Coronavirus Disease 2019 (COVID-19)
Address for correspondence: Anchit Raj Singh, MD, Department of Internal Medicine, Armed Forces Medical College, Solapur Road, Pune, Maharashtra, 411040, India (e-mail: anc.umars@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Context
Due to the wide spectrum of clinical illness in coronavirus disease 2019 (COVID-19) patients, it is important to stratify patients into severe and nonsevere categories. Neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) have been evaluated rapidly by a few studies worldwide for its association with severe disease, but practically none have been conducted in the Indian population. This study was undertaken to examine the role of NLR and PLR in predicting severe disease in Indian patients.
Objectives
The objective was to study the association of NLR and PLR observed at the time of admission with maximum disease severity during hospitalization and to study their role in predicting disease severity.
Material and Methods
A total of 229 COVID-19 patients were admitted at the center during the study period. After applying inclusion and exclusion criteria, 191 patients were included in the study. The demographic, clinical, and laboratory (complete blood count, NLR, and PLR) data of all patients were obtained at the time of admission. Maximum disease severity of all patients was assessed during hospitalization.
Statistical Analysis
Chi-square and Mann–Whitney U tests were used to assess statistical significance. Receiver operating characteristic curve (ROC) was plotted for NLR and PLR to estimate the cutoff values and sensitivity and specificity using Youden's index for predicting severe disease. Logistic regression analysis was used to estimate the odds ratios (OR) and 95% confidence intervals.
Results
Mean NLR and PLR were significantly higher in severe patients (NLR = 7.41; PLR = 204) compared with nonsevere patients (NLR = 3.30; PLR = 121). ROC analysis showed that NLR, in comparison to PLR, had a higher area under the curve (AUC) of 0.779, with a larger OR of 1.237 and cutoff of 4.1, and showed 69% sensitivity and 78% specificity in predicting severe disease. Cut off for PLR was 115.3, which showed 79% sensitivity and 62% specificity in predicting severe disease.
Conclusion
NLR and PLR, both showing acceptable AUCs, can be used as screening tools to predict disease severity. However, NLR was a better predictor of disease severity.
Keywords
COVID-19
hematology
SARS-CoV-2
Introduction
Wuhan city of Hubei Province, China, reported a cluster of cases of atypical pneumonia with unknown etiology in December 2019.[1] Subsequent evaluation led to identification of a “novel” coronavirus that had caused the disease.[2] This new virus was later named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the disease caused by it was named as coronavirus disease 2019 (COVID-19).[3] In India, the first COVID-19 case was reported on January 30, 2020, and more than 3.68 million positive cases had been reported by the end of August 2020.[4]
Due to the increasing patient load and the wide spectrum of clinical illness ranging from asymptomatic infection to fulminant disease, it is imperative to stratify COVID-19 patients based on their severity. Severe patients are few, but they have a longer duration of hospitalization and require ventilatory support. Risk factors have been studied for predicting disease severity in COVID-19 and include age, sex, known comorbidities, biochemical tests such as C-reactive protein, D-dimer, ferritin, procalcitonin, and lactate dehydrogenase, and hematological parameters such as platelet count (PLT), lymphocyte count and neutrophil count.
Hematological parameters are some of the essential laboratory tests that all patients with COVID-19 undergo, regardless of severity. Nevertheless, hematological parameters have been shown to be associated with disease severity. Studies have shown that in nonsevere COVID-19 cases, hematological parameters may be essentially normal, and the severe or fatal cases may show derangements such as leukocytosis, neutrophilia, lymphopenia, eosinopenia, monocytopenia, and thrombocytopenia.[5,6,7,8] Mechanisms underlying these abnormalities include direct effects of viral infection on hematopoietic stem cell differentiation, survival, proliferation, and homing. Indirect effects of COVID-19 are equally important and include effects of inflammatory mediators such as interleukin (IL)-6, which is elevated in severe disease, which affects hematopoietic stem cell proliferation and differentiation. This has generated great interest in studying the utility of hematological parameters and indices as biomarkers that may predict severity.[9]
Using the complete blood counts, many calculated hematological parameters such as neutrophil-to-lymphocyte ratio (NLR), derived NLR, lymphocyte to-monocyte ratio, and platelet-to-lymphocyte ratio (PLR), have been studied in COVID-19. The important hematological parameters that have shown promise in assessing the disease severity and prognostication are NLR and PLR. NLR and PLR have been found to be elevated in severe COVID-19 patients.[9-12] NLR has also been found to be an independent risk factor for mortality of COVID-19 patients admitted to the hospital.[13]
As neutrophils are the first line of defense, they get activated and migrate from peripheral circulation to immune organs in cases of viral infections. Release of reactive oxygen species from neutrophils helps antibody-dependent cell-mediated immunity in killing the virus. Inflammatory factors such as IL-6, IL-8, and tumor necrosis factor-α, which are released by lymphocytes and endothelial cells, activate neutrophils. However, lymphocytes are the mainstay of human immune response in viral infections.[9] Lymphopenia is manifested because the virus may directly infect lymphocytes as these cells express ACE2 (angiotensin-converting enzyme 2) receptors on their surfaces. Furthermore, cytokine storm may cause lymphocyte apoptosis due to increased levels of interleukins. Direct destruction of lymphoid organs such as spleen and thymus may also lead to reduced lymphocyte count. Elevated lactic acid levels, seen in severe COVID-19 cases, might also suppress lymphocyte proliferation.[14] These mechanisms cause increase in NLR with severe disease.
Platelets also play an important role in inflammation. Thrombocytopenia is a common hematological change seen in COVID-19, which can occur due to reduced production of platelets because of direct attack on hematopoietic cells or bone marrow stromal cells by the virus or because of cytokine storm. The lung injury in COVID-19 also can lead to thrombocytopenia by aggregation and retention of platelets in lungs, and the formation of thrombus, causing platelet consumption.[10,15] Platelets also recruit other inflammatory cells to the site of injury after getting activated in response to proinflammatory cytokines. Platelet factor 4, which is released from α granules of activated platelets during platelet aggregation, prevents agglutinin-A from inhibiting lymphocyte formation. Activated platelets also lead to adhesion of lymphocytes to the endothelial cells, causing increased concentration of lymphocytes at the inflammatory site.[10] All these processes contribute to increased PLR.
NLR and PLR have been evaluated rapidly by quite a few studies worldwide in the short time since the origin of COVID-19, but only a few robust studies exist, and practically none have been conducted in the Indian population. This study aims to examine the role of NLR and PLR in Indian COVID-19 patients and their role as predictors of disease severity. The objective was to study the association of NLR and PLR observed at the time of admission with the maximum disease severity during hospitalization and to study their role in predicting disease severity.
Materials and Methods
In this single-center cross-sectional comparative study, we admitted 229 laboratory-confirmed (RT-PCR [reverse transcription polymerase chain reaction] or rapid antigen test) COVID-19 patients at a dedicated COVID hospital in Delhi from July 13, 2010, to August 6. 2020. The protocols for diagnosis and management of COVID-19 was in accordance with the current Ministry of Health and Family Welfare (MOHFW), Government of India (GOI), guidelines.[16] All adult patients were included in the study. Patients with pregnancy, chronic lung diseases, hematolymphoid malignancies, chronic liver disease, blood culture growth showing secondary infection, and malignancy were excluded (►Fig. 1).
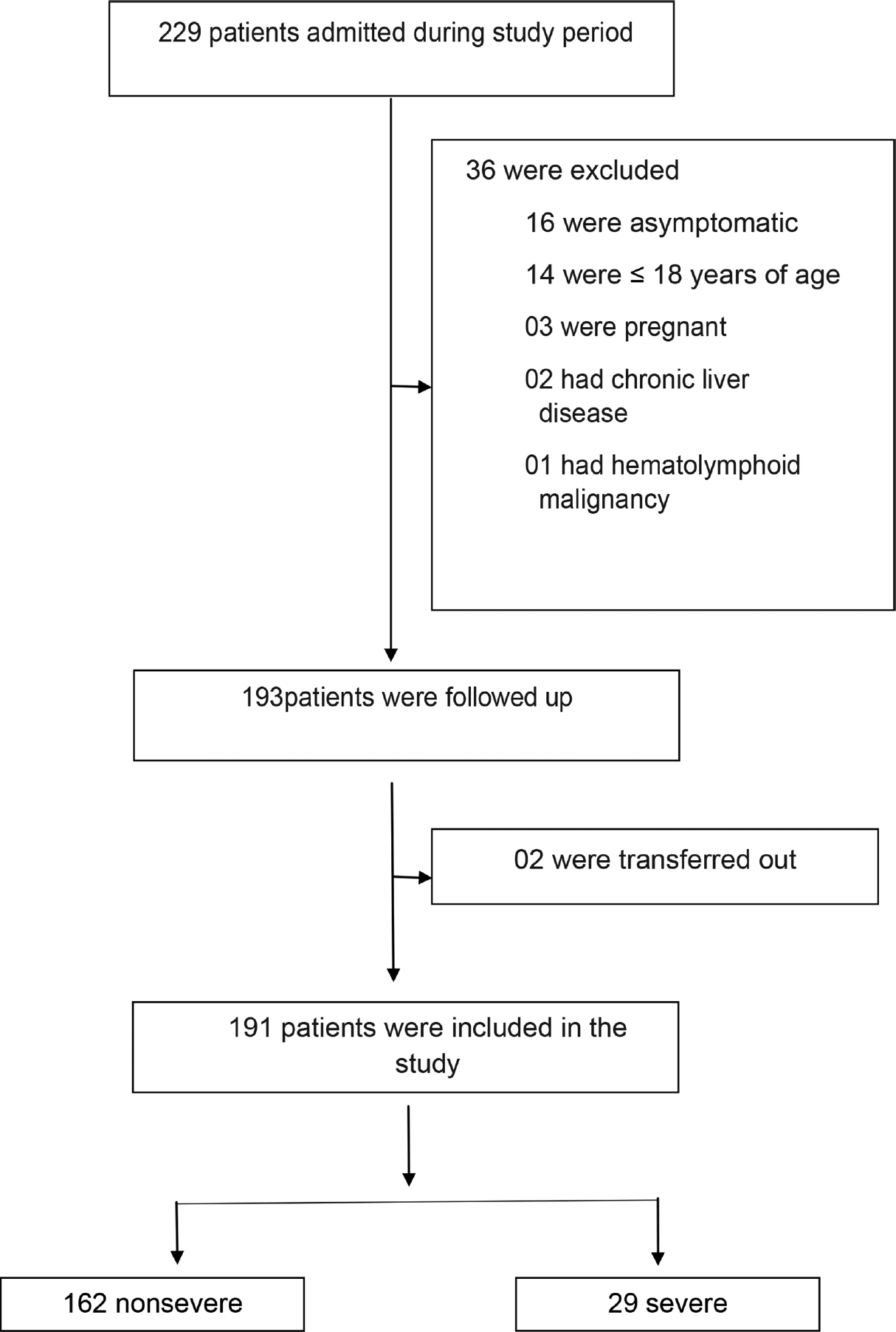
- Flow chart showing enrolment of patients for study.
Clinical data of each patient were collected, which included age, gender, and known comorbidities. History of comorbidities was taken from each patient, which included diabetes mellitus (DM), hypertension (HTN), cardiac diseases, hypothyroidism, chronic kidney disease, and chronic obstructive pulmonary disease. Patient with any one of these diseases was labeled as “patient with comorbidity” (n = 78). Patient with none of these diseases was labeled as “patient without comorbidity” (n = 113). Clinical examination details such as respiratory rate (RR) and saturation of peripheral oxygen (SpO2) were noted for each patient, which were used to assess disease severity as per the latest MOHFW, GOI, guidelines. Based on these parameters, patients were classified into severe cases (RR > 30/minute and/or SpO2 < 90% at room air) and nonsevere cases (RR ≤30/minute and/or SpO2 ≥90% at room air). Each patient's clinical condition was assessed during the stay at the hospital, and maximum severity was noted. The outcome was defined as death or discharge as per the MOHFW, GOI, guidelines.
Informed written consent was obtained from each participant. At the time of admission, 3 mL of venous blood sample was collected into vacutainers containing ethylenediaminetetraacetic acid (EDTA) under aseptic precautions. After calibration and performing internal quality control checks, the blood samples were analyzed using a five-part automated hematology analyzer (Merilyzer CelQuant 5, Meril Life, Gujarat, India), and the following findings were noted: hemoglobin level (Hb), total leucocyte count (TLC), absolute neutrophil count (ANC), absolute lymphocyte count (ALC), and PLT. NLR was calculated by dividing ANC by ALC, and PLR was calculated by dividing PLT by ALC. The interpretation of all hematological parameters was done as per the international standards.[17] Low Hb was considered when Hb was less than 13 g/dL in males and less than 12 g/dL in females. TLC < 4,000 cells/mm3 was defined as leucopenia, and TLC > 10,000 cells/mm3 was defined as leukocytosis. ANC < 1,500 cells/mm3 was defined as neutropenia, and ANC > 7,000 cells/mm3 was defined as neutrophilia. ALC < 1,000 cells/mm3 was defined as lymphopenia, and ALC > 3,000 cells/mm3 was defined as lymphocytosis. PLT < 150,000 platelets/mm3 was defined as thrombocytopenia, and PLT > 450,000 platelets/mm3 was defined as thrombocytosis.
The data were collated in Microsoft Excel and analyzed using SPSS (v. 25, IBM Corp.). Categorical variables were summarized as the counts and percentages in each category. Continuous variables were expressed as the appropriate means and standard deviations. Chi-square and Mann–Whitney U tests were used to assess the statistical significance of difference between groups. p-Value < 0.05 was considered statistically significant. Receiver operating characteristic curve (ROC) was plotted for NLR and PLR, which was used to estimate the cutoff values and sensitivity and specificity using Youden's index for predicting severe disease. Logistic regression analysis was used to estimate the odds ratios (ORs) and 95% confidence intervals. The study protocol was reviewed and approved by the Institutional Ethical Committee.
Results
From the total of 229 patients who were admitted at the hospital during the mentioned period, 191 patients were selected for the study after applying the inclusion and exclusion criteria. Out of these 191 patients, 162 (84.82%) patients were nonsevere and 29 (15.18%) were severe. Four patients (2.09%) died, with 75% cases being ≥ 80 years of age. The remaining 187 patients recovered from the disease. The demographic, clinical, and hematological parameters of these patients are listed in ►Table 1.
| Total (n = 191) | Nonsevere (n = 162) | Severe (n = 29) | p-Value | |
|---|---|---|---|---|
| Age (year), mean ± SD | 44 ± 16.5 | 41.1 ± 15.3 | 59.9 ± 13.8 | < 0.001 |
| Sex (male), n (%) | 140 (73.29%) | 117 (72.22%) | 23 (79.31%) | 0.427 |
| Comorbidities | ||||
| DM, n (%) | 48 (25.13%) | 32 (19.75%) | 16 (55.17%) | 0.001 |
| HTN, n (%) | 53 (27.74%) | 39 (24.07%) | 14 (48.27%) | 0.007 |
| Cardiac disease, n (%) | 15 (7.85%) | 12 (7.41%) | 3 (10.34%) | 0.588 |
| COPD, n (%) | 4 (2.09%) | 3 (1.85%) | 1 (3.45%) | 0.580 |
| CKD, n (%) | 2 (1.05%) | 1 (0.62%) | 1 (3.45%) | 0.168 |
| Hematological parameters | ||||
| Hemoglobin (g/dL), mean ± SD | 12.5 ± 1.82 | 12.7 ±1.79 | 11.6 ± 1.67 | 0.001 |
| TLC (cells/mm3), mean ± SD | 6,717 ± 2,910 | 6,416 ± 2,438 | 8,398 ± 4,454 | 0.016 |
| ANC (cells/mm3), mean ± SD | 4,549 ± 2,648 | 4,196 ± 2,113 | 6,519 ± 4,141 | < 0.001 |
| ALC (cells/mm3), mean ± SD | 1,518 ± 756 | 1,568 ± 719 | 1,237 ± 897 | 0.002 |
| PLT (Lac/mm3), mean ± SD | 1.64 ± 0.831 | 1.59 ± 0.738 | 1.92 ± 1.21 | 0.048 |
| Outcome | ||||
| Recovered, n (%) | 187 (97.91%) | 162 (100%) | 25 (86.21%) | |
| Died, n (%) | 4 (2.09%) | 0 (0%) | 4 (13.79%) | |
Abbreviations: ALC, absolute lymphocyte count; ANC, absolute neutrophil count; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; DM, diabetes mellitus; HTN, hypertension; PLT, platelet count; SD, standard deviation; TLC, total leucocyte count.
The common hematological findings seen in Indian COVID-19 patients were as follows: low Hb ([males, n = 68; 48.57%] and [females, n = 29; 56.86%]), thrombocytopenia (n = 92; 48.17%), lymphopenia (n = 52; 27.22%), leukopenia (n = 24; 12.56%), neutrophilia (n = 23; 12.04%), and leukocytosis (n = 19; 9.9%). These findings were even more common in severe patients showing thrombocytopenia (n = 16; 55.17%), lymphopenia (n = 15; 51.72%), leukocytosis (n = 8; 27.59%), neutrophilia (n = 8; 27.59%), and low Hb ([males, n = 19; 82.61%] and [females, n = 5; 83.3%]).
Mean NLR was significantly higher in severe patients (7.41) compared with nonsevere patients (3.30) (p < 0.001) (►Table 2). Similarly, Mean PLR was also significantly higher in severe patients (204) compared with non-severe (121) patients (p < 0.001) (►Table 2). It was also observed that NLR was higher in patients with comorbidities compared with patients with no comorbidities of similar disease severity (p = 0.72). A similar trend was noted in PLR as well (p = 0.08).
| Total (n = 191) | Nonsevere (n = 162) | Severe (n = 29) | p-Value | |
|---|---|---|---|---|
| NLR, mean ± SD | 3.92 ± 4.35 | 3.30 ± 2.71 | 7.41 ± 8.44 | < 0.001 |
| NLR in patients without comorbidities, mean ± SD | 3.29 ± 2.60 | 3.21 ± 2.63 | 4.59 ± 1.59 | 0.015 |
| NLR in patients with comorbidities, mean ± SD | 4.83 ± 5.95 | 3.46 ± 2.88 | 8.31 ± 9.52 | < 0.001 |
| PLR, mean ± SD | 134 ± 107.0 | 121 ± 80.9 | 204 ± 185.0 | < 0.001 |
| PLR in patients without comorbidities, mean ± SD | 118 ± 71.0 | 117 ± 71.8 | 131 ± 60.8 | 0.222 |
| PLR in patients with comorbidities, mean ± SD | 157 ± 142.0 | 130 ± 95.9 | 227 ± 206.0 | 0.010 |
Abbreviations: NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; SD, standard deviation.
Area under the curve (AUC) for NLR and PLR in the ROC curve analysis was 0.779 and 0.688, respectively (►Table 3 and ►Fig. 2). Furthermore, we estimated the cutoff values of NLR and PLR for differentiating between severe and nonsevere COVID-19 patients, which were 4.1 and 115.3, respectively (►Table 3). Of the two ratios, NLR cutoff of 4.1 had the higher specificity (78%) to differentiate severe and nonsevere patients; however, PLR cutoff of 115.3 was more sensitive (79%). Calculation of OR showed significant odds of severe disease associated with both NLR (1.237; p = 0.001) and PLR (1.005; p = 0.002) (►Table 3).
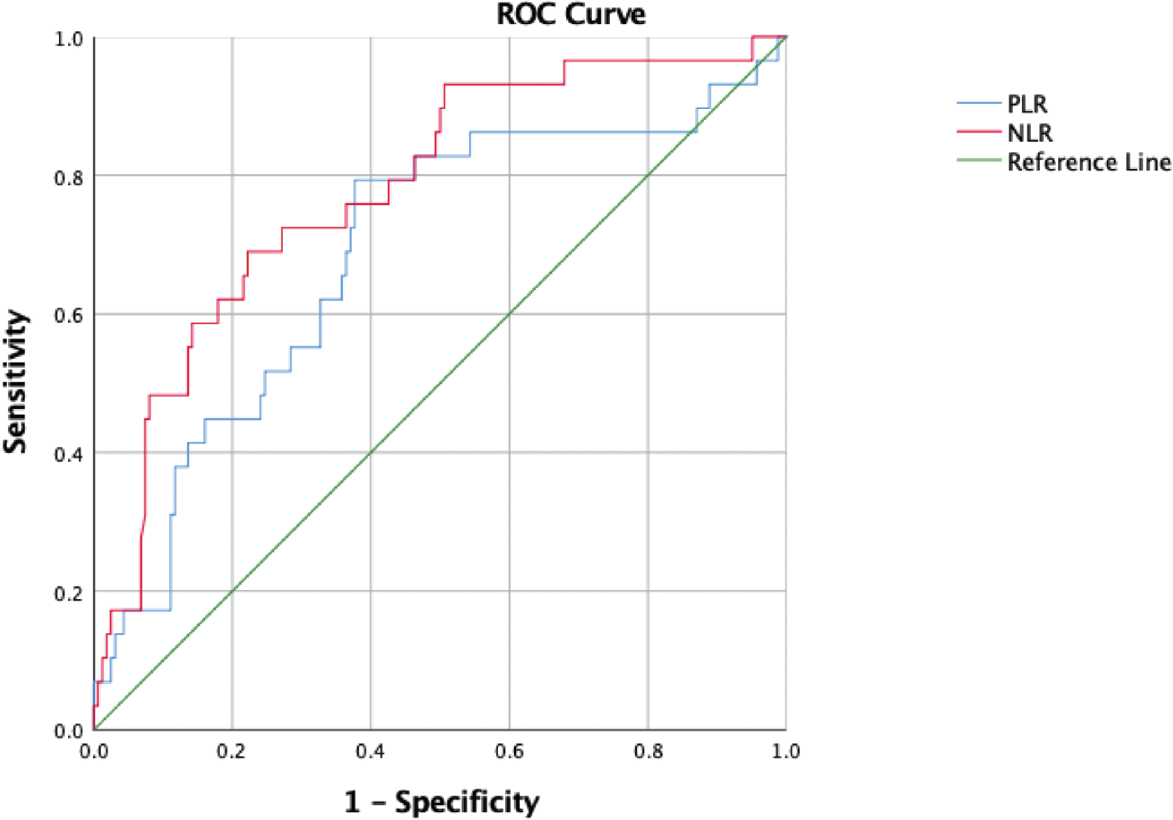
- ROC curve of NLR and PLR to predict the severity of COVID-19. COVID-19, coronavirus disease 2019; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; ROC, receiver operating characteristic curve.
| Parameter | AUC | Cutoff value | Sensitivity (%) | Specificity (%) | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| NLR | 0.779 | 4.1 | 69 | 78 | 1.237 (1.090–1.405) | 0.001 |
| PLR | 0.688 | 115.3 | 79 | 62 | 1.005 (1.002–1.009) | 0.002 |
Abbreviations: AUC, area under the curve; CI, confidence interval; COVID-19, coronavirus disease 2019; NLR, neutrophil-to-lymphocyte ratio; OR, odds ratio; PLR, platelet-to-lymphocyte ratio.
Discussion
Common hematological changes seen in COVID-19 patients were thrombocytopenia, lymphopenia, neutrophilia, and leukocytosis, which are consistent with previous studies.[5-8] Our study also showed that NLR and PLR were higher in severe patients than nonsevere, which is also similar to previous reports.[9,11,12] This study also suggested that NLR and PLR with acceptable AUCs of 0.779 and 0.668, respectively, can be used as screening tools to predict disease severity. Out of these two, NLR was found to be a better predictor of disease severity with a numerically larger OR revealing a larger effect size.
The distribution of comorbidities, such as DM and HTN, and advanced age were significantly higher in the severe group, owing to them being independent risk factors for severe disease. However, our study showed that there was no significant difference in NLR and PLR with comorbidities. This study was a hospital-based study and did not include asymptomatic patients of COVID-19; therefore, generalizability of results to a community setting may not be possible, and this might be a limitation of the study.
To summarize, NLR and PLR can be helpful in predicting COVID-19 severity in our country, especially in resource-limited settings where sophisticated biochemical markers might not be easily available. They are also cheap and easy to calculate. Further studies may be needed to improve their diagnostic accuracy by combining them with other parameters that may improve their diagnostic accuracy. They may help clinicians in evaluation and risk stratification of COVID-19 patients.
Conclusion
In conclusion, we found out that NLR and PLR were significantly higher in severe COVID-19 patients than nonsevere patients. With the use of ROC curve, we provided the cutoff for these parameters, which can classify the patients into severe and nonsevere categories. This cutoff for NLR was 4.1 and that for PLR was 115.3.
Author Contributions
R. J. and A. G. designed the study, collected the data, and prepared the manuscript. R. J., A. G., A. S., and B. P. contributed in data and statistical analyses. S. M. and T. T. were involved in manuscript editing and review. All authors had full access to the data and vouch for the accuracy of the analyses. All authors approved the final manuscript and supported the decision to submit for publication.
Permission
Permission from the Institutional Ethical Committee was taken prior to the study.
Acknowledgments
We thank Mr. Amit Jain for valuable comments and support regarding data analysis.
Conflict of Interest
None.
References
- Pneumonia of cause – China. Available at: http://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/. Accessed July 7, 2020
- [Google Scholar]
- New-type coronavirus causes pneumonia in Wuhan. Available at: http://www.xinhuanet.com/english/2020-01/09/c_138690570.htm. Accessed July 7, 2020
- [Google Scholar]
- China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China,N Engl J Med. 20192020;382(08):727-733.
- [CrossRef] [PubMed] [Google Scholar]
- Coronavirus Cases. Available at: https://www.worldometers.info/coronavirus/country/india/. Accessed August 31, 2020
- [Google Scholar]
- Coronavirus disease 2019 (COVID-19): a short review on hematological manifestations. Pathogens. 2020;9(06):493.
- [CrossRef] [PubMed] [Google Scholar]
- Hematological findings and complications of COVID-19. Am J Hematol. 2020;95(07):834-847.
- [CrossRef] [PubMed] [Google Scholar]
- Hematological features of persons with COVID-19. Leukemia. 2020;34(08):2163-2172.
- [CrossRef] [PubMed] [Google Scholar]
- Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58(07):1021-1028.
- [CrossRef] [PubMed] [Google Scholar]
- The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84 106504
- [CrossRef] [PubMed] [Google Scholar]
- Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease-19. J Med Virol 2020 (e-pub ahead of print) DOI: 10.1002/jmv.25767
- [CrossRef] [PubMed] [Google Scholar]
- The value of clinical parameters in predicting the severity of COVID-19. J Med Virol 2020 (e-pub ahead of print) DOI: 10.1002/jmv.26031
- [CrossRef] [PubMed] [Google Scholar]
- Use of neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios in COVID-19. J Clin Med Res. 2020;12(07):448-453.
- [CrossRef] [PubMed] [Google Scholar]
- Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect. 2020;81(01):e6-e12.
- [CrossRef] [PubMed] [Google Scholar]
- Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5(01):33.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanism of thrombocytopenia in COVID-19 patients. Ann Hematol. 2020;99(06):1205-1208.
- [CrossRef] [PubMed] [Google Scholar]
- COVID19, Ministry of Health and Family Welfare (MOHFW), Government of India (GOI) dated. 03/07/2020; Version 5
- [Google Scholar]





